Determination of X-ray Crystal Structures of ERAP1 Complexes
We were asked by Grey Wolf Therapeutics (in collaboration with Sygnature Discovery) to provide support for an endoplasmic reticulum aminopeptidase-1 (ERAP1) drug discovery program. In addition to providing purified ERAP1 proteins for assay, we were also asked to determined X-ray crystal structures of ERAP1 in complex with a number of project compounds.
Grey Wolf Therapeutics is a UK-based drug discovery biotechnology company focused on immuno-oncology. Their approach is to directly alter the tumour cells, increasing tumour visibility and extending the reach of immunotherapy in oncology. Grey Wolf’s in vitro and in vivo non-clinical studies have demonstrated that targeting ERAP1 in the antigen presentation pathway drives modulation of the neoantigen repertoire on tumour cells.
ERAP1 belongs, together with ERAP2 and insulin-regulated aminopeptidase, to the oxytocinase subfamily of M1 zinc-metallopeptidases. Originally described as an aminopeptidase with activity toward peptide hormones (e.g. angiotensin II), it was subsequently reported as a critical component of the final processing of a discrete but wide repertoire of MHC-I ligands in the ER.
Genome-wide association studies have revealed that ERAP1 is a susceptibility factor for ankylosing spondylitis (AS) and other MHC-I associated diseases. Evidence has also been accumulating that ERAP1 plays a significant role in tumour control and immune-surveillance. ERAP1 is therefore an attractive target for the development of small molecule inhibitors in order to treat a range of diseases.
At the time, only three published crystal structures of ERAP1 were available in the PDB and all were at medium resolution (2.7-3.0 Ång). One of the structures was of the closed or active form in complex with the protease inhibitor bestatin which fortuitously bound during purification (PDB code: 2YD0) while the other two were in open or inactive conformations (3QNF, 3MDJ). Protein for all three of these literature structures was produced in baculovirus/insect cells and was N-glycosylated at three sites.
These X-ray structures revealed that ERAP1 consists of 4 domains (I–IV) (Fig. 1). Domain I is located on top of the catalytic domain II and includes residues required for binding the N-termini of substrate peptides. The small domain III has a β-sandwich structure while domain IV is a large α-helical domain, which extends away from the catalytic site and forms a large cavity which, together with domain II, is the main substrate binding site. This cavity is relatively shallow near the active site but widens substantially in domain IV.
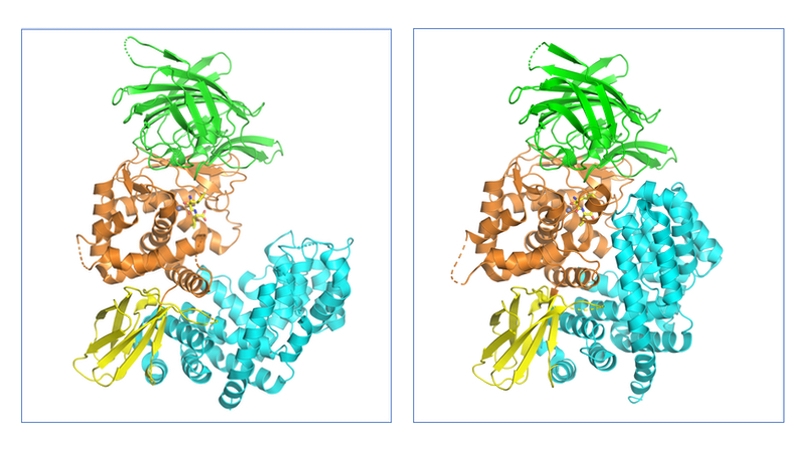
Figure 1. Structures of the open/inactive (PDB code: 3MDJ) and closed/active (PDB code: 2YD0) forms of human ERAP1. The four domains of ERAP1 and coloured separately: domain I (green), domain II (orange), domain III (yellow) and domain IV (teal).
The large conformational change between the apo (open) and substrate bound (closed) states was observed to be due to a rotation of domain III, hinging on the interdomain region between domains II and III region, so that domain IV is pulled in toward domain I (Fig 1.).
One of the main objectives of the work was to improve on the resolution of the then published X-ray structures so they could be a better guide to chemistry design.
We were able to show early in the project that protein yields in mammalian cells were significantly higher than that in insect cells. Furthermore, the protein could be produced with most of the glycosylation removed by kifunensine/endoH treatment which leaves only a single GlcNAc on the glycosylated asparagines.
While our initial crystallisation experiments generated large crystals, their diffraction was still considered insufficient for structure-based ligand design. Therefore, in order to improve the resolution of the structures further, a two-pronged strategy was utilised (i) construct engineering and (ii) buffer screen using a DSF thermal shift assay.
ERAP1 constructs were designed that deleted a number of residues near the interdomain II–III hinge region that were observed to be disordered in a number of the published structures.
The DSF buffer screen revealed that there was a significant thermal stabilisation with particular buffer conditions and that both deletion of hinge residues and binding of selected project tool compounds also showed a clear improvement in thermal stability.
Utilising this information, we were quickly able to obtain ERAP1 crystals in complex with both known tool inhibitors (Fig. 2) as well as the client’s proprietary compounds that diffracted to 1.6-1.7 Ång resolution. This was a substantial improvement in resolution on published structures at the time. These structures enabled confirmation of binding mode of existing project lead compounds, rationalisation of SAR data and subsequent ligand design.
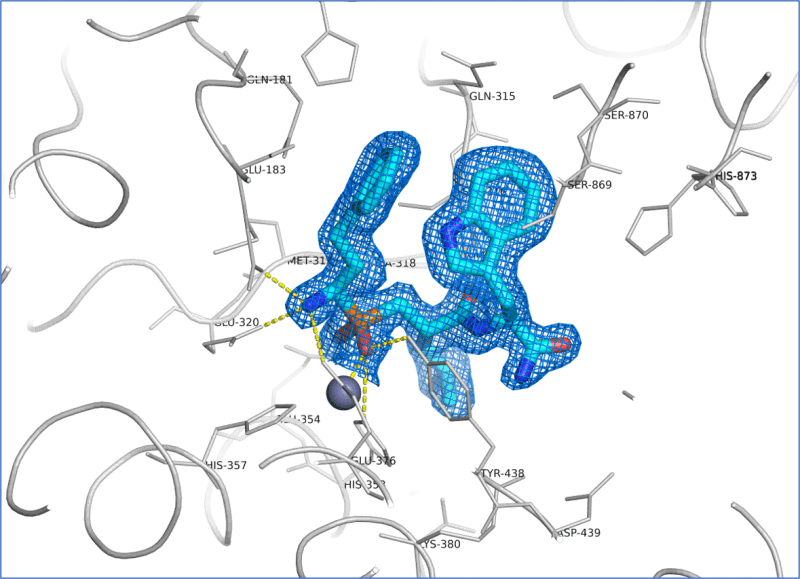
Figure 2. Binding pocket of ERAP1 containing the tool compound ligand DG013A solved at 1.66 Å resolution. The ligand molecule (cyan) is shown superimposed with the refined 2Fo-Fc electron density map contoured at 1.0 σ. Hydrogen bonds are indicated as yellow dotted lines.
“Developing structural data for our ERAP1 program has been an important part in enabling Grey Wolf to focus the lead optimisation efforts and drive a greater understanding of the key interactions our lead molecules make with the protein. Peak Proteins have not only provided the program with multiple protein isoforms for enzyme and biophysical assays across our screening cascade but tackled the crystallography work from several angles to ensure success”
Dr Peter Joyce
Chief Executive Officer and Co-founder, Grey Wolf Therapeutics


