Preventing Protein Aggregation to Aid Crystallisation
The final purification step prior to protein crystallisation is frequently size-exclusion chromatography in the desired buffer. This enables the purification of a single oligomeric species making successful crystallisation much more likely. A frequent challenge whilst purifying proteins however is their tendency to aggregate. This propensity to aggregate varies quite considerably between proteins. There are a number of reasons for this which can include the miss-folding of the protein to expose the hydrophobic core or the protein surface itself being unusually hydrophobic.
Our Solution
We therefore decided to see if it was possible to engineer a version that was less prone to aggregation and more straightforward to purify. We aligned the sequence of our target protein with the sequences of the species orthologues whose structures had been solved. By doing this and analysing the structures themselves we were able to identify five very hydrophobic residues in our target protein that were generally aliphatic in the other sequences and therefore likely to be on the surface. These were all mutated to alanine residues. This protein expressed well and could be purified by Ni-affinity in a manner similar to before. However, it was now significantly easier to remove the 6His-tag by treatment with TEV protease and the cleaved protein was readily separated from uncleaved and other contaminants on a second Ni column. More importantly when purified by size-exclusion chromatography the protein was now mostly monomeric in nature. After removal of detergent the protein gave a very clear and expected intact mass and generated a very clear thermal melting curve indicating that it was nicely folded.
SEC chromatograms of wild type (above top) and the mutated construct (above bottom) showing the ratio of aggregate to monomer has been reversed, and the monomer peak at 11ml has become the larger component in the protein sample for the engineered construct.
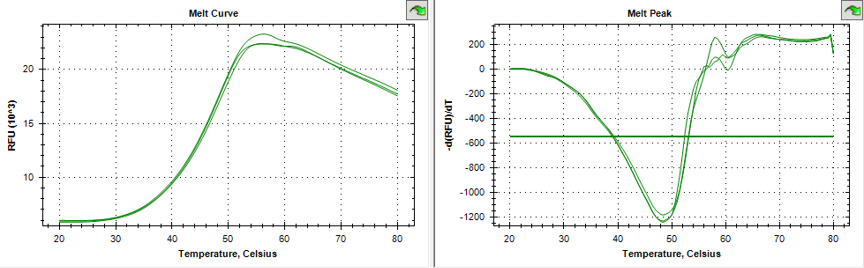
Triplicate melting curves of protein using Thermofluor . Midpoint of the unfolding transition is 48.5°C, showing a stable, well-folded protein.
The Impact
We are now able to much more confidently take the protein into crystallisation experiments without concerns over the quality of the protein. These experiments are currently ongoing.


