Removing N-linked Oligosaccharides to Aid Protein Crystallisation
A client was looking to solve the crystal structure of a secreted protein which contained a number of disulphide bonds and potential N-linked glycosylation sites, which leads to the attachment of complex sugar molecules. There was already literature available describing the expression of the protein in an E.coli strain (proteins do not get N-glycosylated in E.coli) that supports disulphide bond formation, but this was proving difficult to reproduce, with almost all the protein being expressed in an insoluble form. We suggested an alternative approach to secrete the protein from a transient HEK cell system where the disulphide bonds should form correctly, but also would lead to glycosylation of the protein. Being large, bulky and of differing sizes, added sugar molecules can affect a protein’s ability to form uniform crystals. Therefore, any N-linked complex sugars added would need to be removed before crystallisation.
Our Solution
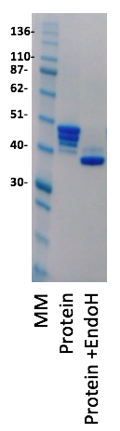
The protein was very well expressed from the HEK cells as predicted and could be easily purified using a combination of Ni affinity and size exclusion chromatography. The fact that HEK cells grow in a protein free media also made purification much more simple. What was clear from the SDS-PAGE analysis was that the protein had a number of different glycosylated forms with 1, 2, 3 and 4 of the N-linked sites being glycosylated. Our approach to removing the problematic sugars was to grow the cells in the presence of the mannosidase inhibitor, kifunensine, which results in an oligosaccharide structure that is sensitive to endoglycosidase H (EndoH) cleavage of the oligosaccharide (Chang, VT et al., 2007). EndoH treatment of the protein leaves a single N-acetyl glucosamine with a mass of 203Da. When the protein was expressed in the presence of kifunensine, purified and treated with EndoH this resulted in deglycosylation as shown in the SDS-PAGE gel below.
The protein was then analysed using protein mass spectrometry to calculate the intact mass. This revealed a number of species that had a mass +203Da, +406Da, +609Da and +812Da corresponding to N-acetylglucosamine present at 1, 2, 3 or 4 sites on the protein, exactly as expected from the EndoH cleavage of the N-linked structures.
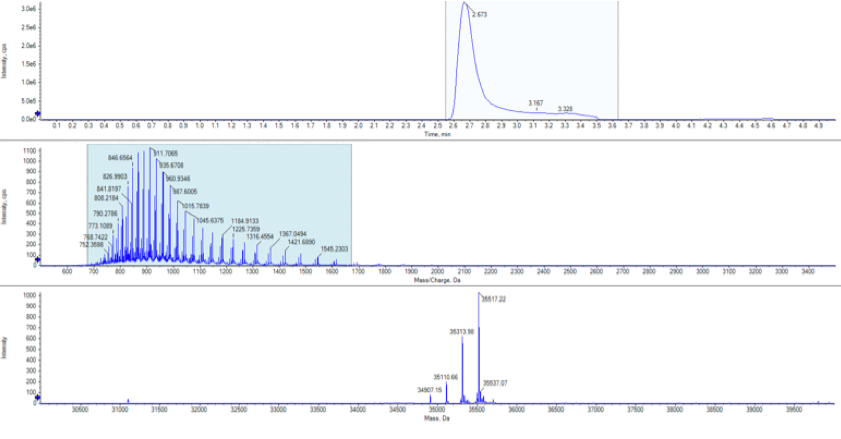
Intact mass was performed on the Sciex X500B system coupled to a Sciex Exion LC
The Impact
Expression of the protein in a secreted form in our HEK cell system, alongside kifunensine and EndoH treatment, provided our client with the correctly folded, deglycosylated protein in a form suitable for crystallisation. Switching from a specific E.coli strain, where it was difficult to reproduce literature findings, to a simple mammalian alternative approach, allowed the project to progress successfully to the next stage.