What is Fragment-Based Lead Generation (FBLG)?
Fragment-based lead generation (FBLG) is a very popular paradigm for early hit identification and lead generation and can be an attractive alternative to the more traditional high-throughput screening (HTS) methods. An interesting example of this was recently published in Nature Chemistry (Mousnier et. al., 2018) in which fragment-based, pico-molar inhibitors of human N-myristoyl transferase (NMT1 & NMT2) were developed in order to block capsid assembly and replication of the common cold virus. By targeting the host enzymes, that are responsible for an essential post-translational modification of a key viral protein, they were able to neatly sidestep the otherwise tricky problems of there being more than 100 different viral serotypes, and the high mutational rate of the virus, which greatly increases the odds of resistance.
The basic concept behind FBLG is to screen a relatively small library (around 1000) of low molecular weight compounds or fragments (typically approx. 200 Da) rather than millions of larger, often highly elaborated, molecules (400-500 Da) that are normally used in high throughput screening. Because they are small, the fragments are much less likely to contain portions that prevent binding and more likely to have functional motifs that match the chemical environment of the target binding sites, thereby sampling available chemical space much more efficiently. This small molecular size however also has the disadvantage that their binding affinity is generally rather low (typically 100uM to a few mM) thus requiring the use of sensitive biophysical assays to detect this weak binding.
 The methods most often used to do this are: ligand observed NMR, surface plasmon resonance(SPR) or X-ray crystallography. Each of these techniques has its own particular advantages. For instance, NMR is a label-free solution method in which the solubility and stability of both the protein and fragment can be easily monitored to help prevent both false negative and positive results. SPR uses smaller amounts of protein and can be used to screen relatively rapidly. Screening by X-ray crystallography often results in a higher hit rate as it’s usually performed with very high fragment concentrations (10-50 mM), although the requirement of having a suitably robust crystal system can be a major hurdle for some targets.
The methods most often used to do this are: ligand observed NMR, surface plasmon resonance(SPR) or X-ray crystallography. Each of these techniques has its own particular advantages. For instance, NMR is a label-free solution method in which the solubility and stability of both the protein and fragment can be easily monitored to help prevent both false negative and positive results. SPR uses smaller amounts of protein and can be used to screen relatively rapidly. Screening by X-ray crystallography often results in a higher hit rate as it’s usually performed with very high fragment concentrations (10-50 mM), although the requirement of having a suitably robust crystal system can be a major hurdle for some targets.
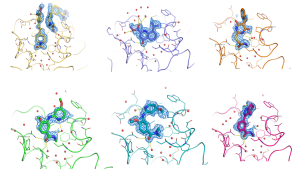
However, even if not used as the primary fragment screening methodology, X-ray crystallography and its ability to generate high-resolution structures of protein-ligand complexes are almost essential for being able to successfully progress the initial fragment hits into lead compounds. For most targets with only weak binding fragments, it is very difficult to generate useful SAR for a fragment by trial and error, as most modifications will result in a loss or change of affinity that is difficult to rationalise without an accurate model of how they are binding to the target. Being able to actually visualise the binding mode of the different fragments and seeing what their interactions are with the protein greatly enables the fragment to lead optimisation process. The great advantage of starting with such small hits is that it can often be much easier to design lead compounds with more favourable physiochemical properties, as affinity is increased, than when starting with the larger more elaborate compounds normally obtained by traditional HTS.
Another such example of fragment screening by SPR followed by X-ray crystallography to visualise the binding modes of the hits can be seen here. Working closely with Sygnature Discovery, who screened their proprietary fragment library against the target BRD3 using SPR, nine confirmed fragment hits were obtained with affinities <250uM. Peak Proteins was then able to solve high-resolution X-ray crystal structures of six of these initial hits shown in the image above, greatly enabling subsequent chemical design and optimisation.


