Beyond N-Glycans: Exploring O-Linked Glycosylation of Proteins
In this article we discuss O-linked glycosylation of proteins and show some data from a study with the platelet receptor G6b-B to exemplify how we approach identifying the sites of sugar attachment.
Glycosylation is a common post-translational modification of proteins in eukaryotic systems. Oligosaccharides attached to asparagine or N-linked structures are very common, extensively studied and engineered for clinical and research uses. They have an obligatory consensus of N-X-S/T where X can be any amino acid except proline. Much less common are oligosaccharides linked to the -OH group of either threonine or serine. These do not have a strict consensus although a bioinformatic tool can be used to predict sites.
At Peak Proteins (Part of Sygnature Discovery) we generate more than 100 different secreted recombinant proteins every year using primarily HEK and CHO cell systems. The majority of these will be N-glycosylated. We will routinely remove the N-linked glycans by either mutating the asparagine or more commonly culturing the cells in kifunensine and then treating the protein with endoglycosidase H 1.
Over a number of years when analysing secreted proteins using intact mass spectrometry, we have very occasionally observed a mass that is 948 Da higher than the expected mass. We first observed this with the platelet receptor G6b-B 2. This mass is consistent with a tetrasaccharide structure of a N-acetyl galactosamine, galactose and two sialic acids O-linked to a Ser or Thr. In the case of G6b-B we were able to demonstrate using mutational studies that the site of glycosylation was T73. Interestingly though we observed that a T73A mutant was still ~ 10-20% O-glycosylated. We never observed a mass increase of 1896 Da and we hypothesised that once one site was O-glycosylated it prevented the modification of additional adjacent sites.
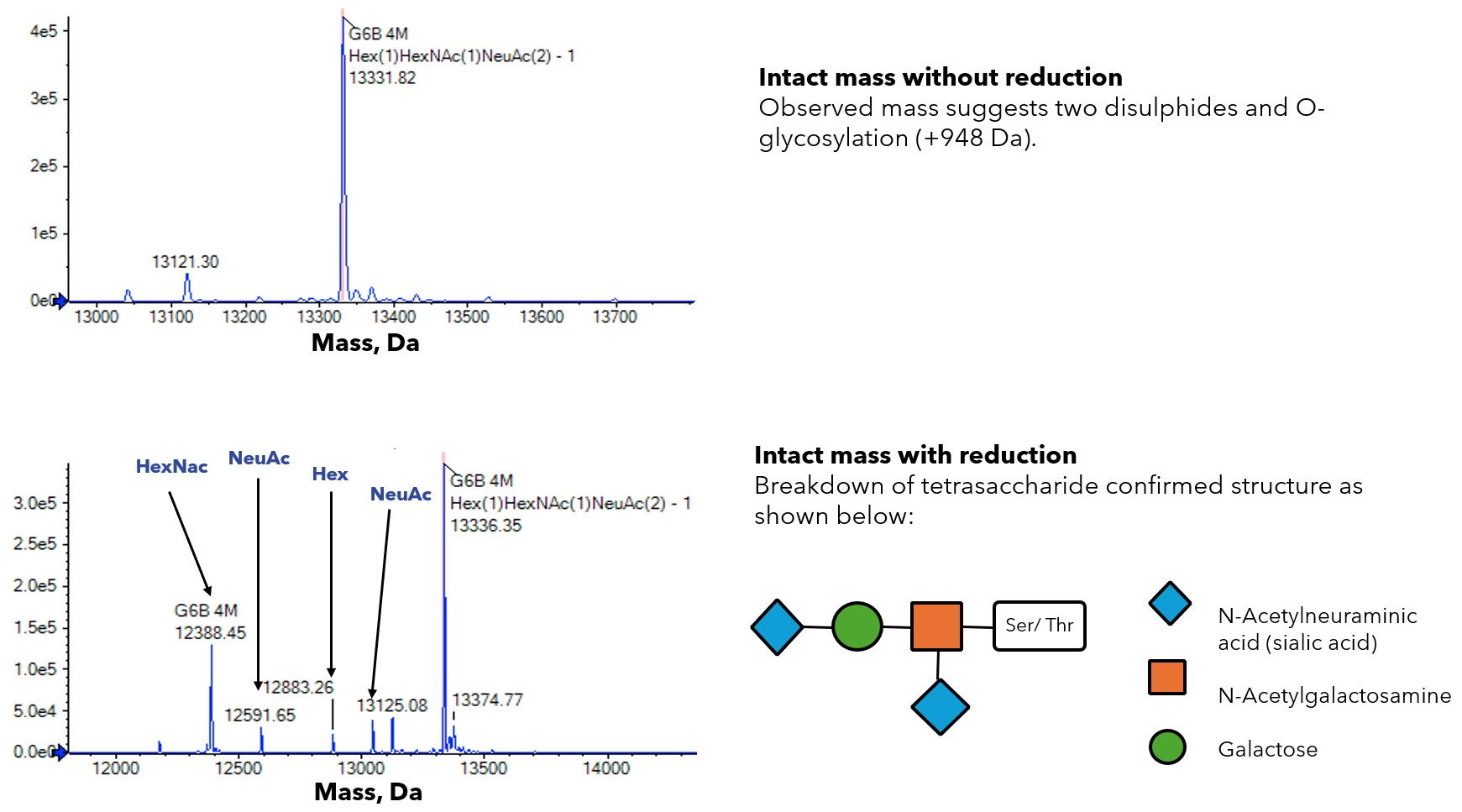
Figure. 1 – Mass Spectrometry analysis of G6b-B
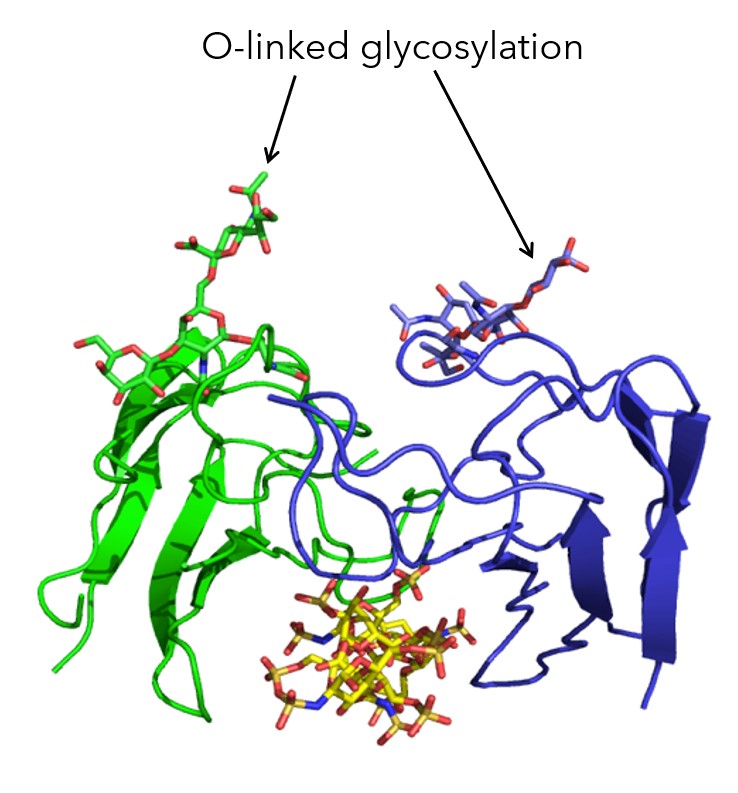
Figure. 2 – Crystal Structure solved at Peak Proteins (Part of Sygnature Discovery) showing the O-linked glycosylation of G6b-B
Since this original observation we have seen a similar mass increase of 948 Da from time to time. We reported one of these in a previous case study. More recently we have observed the same mass increase in a couple of cytokines and a secreted transferase enzyme. We have observed this irrespective of the host being HEK or CHO cells. Unlike G6b-B where a single species of +948 Da was observed, the masses seen on other proteins have been much more heterogeneous. In all three cases there was a mixture of +/- the O-linked structure with heterogeneity of the number of saccharide residues.
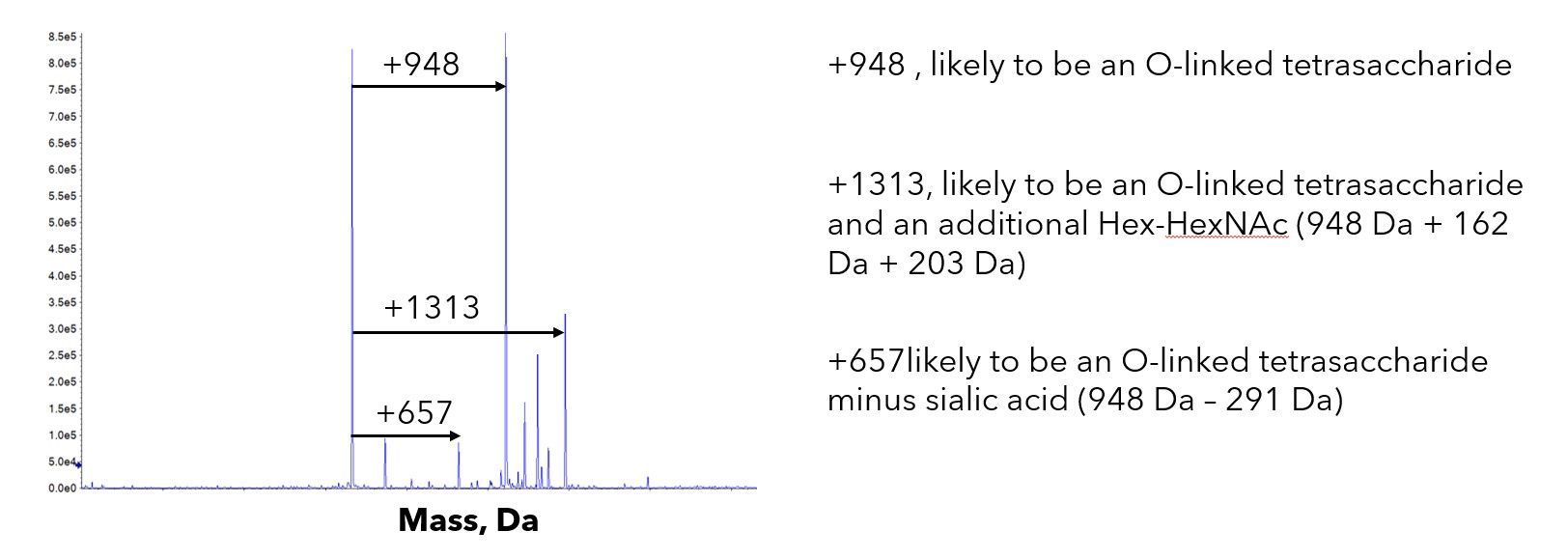
Figure. 3– Heterogeneity of O-linked glycosylation observed in other proteins
Peptide mapping has in all cases suggested that a threonine adjacent to a proline has been modified. Interestingly the secreted transferase had an N-terminal 6His tag followed by a 3C protease (LEVLFQGP) site and then a threonine. It was the N-terminal threonine next to the proline of the 3C site that had been modified. These observations are consistent with the detailed studies we performed with G6b-B where a threonine next to proline is the preferred site.
Interestingly a recent publication has described the O-glycosylation of the stalk region of the protein PD-1 3. In their work using a combination of methods they have identified both threonine and serine modified residues, and in all cases, they are adjacent to proline. Thus, although not a strict consensus as with N-glycosylation there does appear to be an emerging pattern of Thr or Ser next to Pro as a strongly preferred location.
The biological function of these small oligosaccharide structures continues to be investigated. One function appears to be glycan-mediated protection from proteolysis which can play a role in coregulating the release of ectodomains from membrane proteins. An interesting example of this is tumour necrosis factor α (TNFα), which has controlled release via the metalloprotease ADAM17. Regulation of TNFα’s O-glycosylation by GalNAc-T2 plays a role in this release 4,5. There are also structural roles for these glycans, examples include improving solubility through water binding to sialylated O-glycans in mucins 6 and decreasing aggregation in IL-2 7.
O-glycosylation can have a significant impact on proteins produced for biotherapeutic purposes – affecting protein stability and half-life, antibody recognition, protease recognition and target engagement. Here a Peak Proteins (Part of Sygnature Discovery), we have an in-house mass spectrometry service which we use to better understand the proteins we produce and have extensive experience identifying modifications and producing biotherapeutic proteins/antibodies.
References
1. Chang, V. T. et al. Glycoprotein structural genomics: solving the glycosylation problem. Structure 15, 267–273 (2007).
2. Vögtle, T. et al. Heparan sulfates are critical regulators of the inhibitory megakaryocyte-platelet receptor G6B-B. Elife 8, (2019).
3. Tit-oon, P. et al. Intact mass analysis reveals the novel O-linked glycosylation on the stalk region of PD-1 protein. Scientific Reports 2023 13:1 13, 1–12 (2023).
4. Takakura-Yamamoto, R., Yamamoto, S., Fukuda, S. & Kurimoto, M. O-glycosylated species of natural human tumor-necrosis factor-alpha. Eur J Biochem 235, 431–437 (1996).
5. Takakura-Yamamoto, R., Yamamoto, S., Fukuda, S. & Kurimoto, M. O-glycosylated species of natural human tumor-necrosis factor-alpha. Eur J Biochem 235, 431–437 (1996).
6. Crouzier, T. et al. Modulating mucin hydration and lubrication by deglycosylation and polyethylene glycol binding. Wiley Online Library T Crouzier, K Boettcher, AR Geonnotti, NL Kavanaugh, JB Hirsch, K Ribbeck, O Lieleg Advanced Materials Interfaces, 2015 Wiley Online Library 2, (2015).
7. Rosenberg, A. S. Effects of protein aggregates: an immunologic perspective. AAPS J 8, (2006).

