Revolutionising our Protein Science: How FIDA Enhances Protein Projects at Peak Proteins
Written by Dr Duncan Smith and Dr Martin Jennings
At Peak Proteins, we have been utilising the Fida platform (Flow Induced Dispersion Analysis) for the past six months, and it has quickly become an essential tool for our team. Fida has contributed as an analytical tool in a number of our protein projects, as well as enabling stand-alone Fida projects. Here we discuss what Fida is, how Fida has helped us to advance our protein science projects, and how it can benefit our clients. We will also showcase some example data generated at Peak Proteins to illustrate the potential of Fida.
What is Fida?
Fida is a technique which allows the measurement of a biomolecule’s size (typically proteins for us) in solution. This method uses diffusion under laminar flow to mobilise the sample through a capillary, allowing for the calculation of hydrodynamic radius (Rh) from peak width. At Peak Proteins, we can follow proteins by intrinsic or labelled fluorescence giving experimental flexibility. Fida is incredibly versatile and can provide a wealth of information on particle size, oligomeric state, buffer effects, protein stability, aggregation, and protein-ligand interaction. It is relatively quick to perform with a standard run taking 6 minutes, and some types of experiment can use just nanolitres of sample.
Fida for QC Analysis
We regularly use Fida for QC analysis on proteins purified at Peak Proteins. Fida offers a quick homogeneity check (Fig. 1), which is particularly useful for proteins that may be prone to aggregation. We also use Fida to check protein stability over time or a freeze/thaw step, including measuring membrane protein solubility and stability where Fida offers an alternative to techniques such as fluorescent size exclusion chromatography (FSEC; Fig. 2). Fida has been especially useful for large species that are too big for SEC, such as megadalton protein complexes or liposomes, providing a complementary technique to obtain valuable data.

Figure 1. A) SDS-PAGE of 12 subunit protein complex (left) and FIDA analysis (right). B) SEC profile showing 2 populations (top) and FIDA analysis (below). Peak 1 is a soluble oligomer, and Peak 2 is the desired monomeric protein C) Successful labelling of an antibody with Alexafluor 488. Multi-species fit reported two identical sizes (right), indicating a monodisperse sample and no excess free dye.
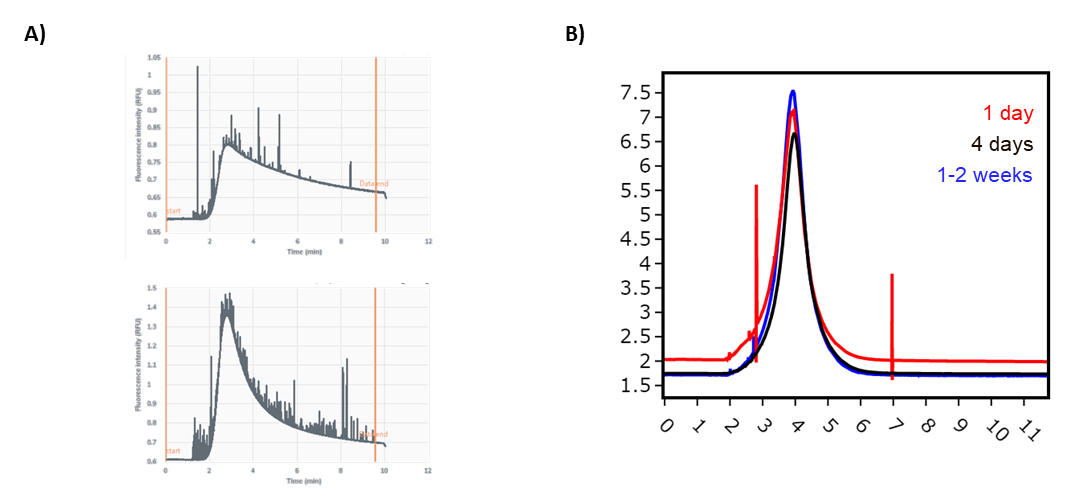
Figure 2. A) Protein instability before (top) and after concentration and dilution (bottom) B) Stability of protein reconstituted into liposomes measured over time.
Measuring Interactions using Fida
Fida is a powerful tool that can be used to measure interactions in solution by monitoring size changes.Kd values can be estimated by measuring the apparent size of the ligand at different analyte concentrations and plotting a binding curve. We have used Fida to measure protein-protein and protein-DNA interactions (Fig. 3).

Figure 3. A) Interaction between Spike-AF488 and ACE2. B) Interaction between protein mutants and FAM labelled DNA.
Measuring Oligomeric State
Fida is an excellent tool for assessing oligomeric state by measuring the absolute size of the analyte and comparing it to expected/predicted sizes. At Peak Proteins, we have used this to measure protein self-affinity and establish monomeric conditions (Fig. 4) for in-house crystallography projects.

Figure 4. Self association between labelled protein (constant) and titrated unlabelled protein
Where Will Fida Take Us Next
Fida has had a significant impact on most areas of our work. It has been applied to dedicated projects and used as a tool to work smarter, sustaining high interest over multiple projects with several clients. One of the most important features of Fida is its quick time to first result, which allows us to rapidly evaluate protein samples and binding interactions. We can usually determine whether an interaction will be suitable for a Fida study within hours. In the next six months, we expect to focus on measuring protein-protein and protein-ligand interactions, studying oligomeric state, and exploring PROTACs – and that’s just scratching the surface of Fida’s capabilities, which include applications such measurement of protein aggregation and liquid-liquid phase separation.
Conclusion
Overall, Fida has proven to be an effective, versatile, and valuable platform for Peak Proteins. It has enabled us to advance our protein science projects and offer new capabilities to our clients. We are excited about the future of Fida and the possibilities it holds for our research and our clients. If you would like to discuss how Fida can help in your projects, please contact us.

