Measuring Coronavirus Spike:ACE2 Binding Affinity and Kinetics Using the Biacore 8K
by Dr. Steven Harborne and Dr. Raquel Faba Rodríguez
Peak Proteins is assisting in the fight against COVID-19 by supplying protein reagents for coronavirus research. For example, we are supplying a research team from Trinity College Dublin to help develop diagnostic tests for antibodies against the virus, or providing reagents to probe the cellular mechanisms of coronavirus infection through a collaboration with the Medicines Discovery Catapult and Retrogenix. The main focus of these studies is the interaction between the viral spike (S) protein and the human angiotensin converting enzyme (ACE2), a critical step in the process of viral infection. Our challenge was to rapidly express and purify these protein components and validate that they have relevant physiological function. All proteins were purified from HEK293 cells as a single species as shown by SDS-PAGE and size exclusion chromatography. Luckily, through the Alderley Park Open Access lab we had access to a leading tool in the measurement of protein:protein interactions: the Biacore 8K from Cytiva.
Remote crash course on the Biacore 8K
Unfortunately, we were complete novices to surface plasmon resonance (SPR) and had never used the Biacore 8K before. Furthermore, due to lockdown and then social distancing it was looking very challenging for us to be trained. Luckily, through the excellent guidance of our local Cytiva technical specialists and reps, Claire Shepherd, Anette Person and Catherine O’Brien, we were provided with a series of virtual presentations and remote support to enable us to get up running in no time!
ACE2 was sticky!
In our initial assay design, we immobilised our Histidine-tagged S protein constructs onto an NTA coated chip, and used varying concentrations of human ACE2 as the analyte. Success! We were able to see clear, specific binding of ACE2 to the immobilised S proteins – but, there was a problem… (Figure 1). We noticed there was non-specific binding in the reference cell. The reference cell itself acts as a control for observing such non-specific interactions, but also allows blank subtraction of resultant sensorgrams, removing fluctuations in baseline due to injections. Binding in the reference cell creates a problem, as subtraction of the reference cell sensorgram caused a downward drift in readings from the active cell, making it impossible for us to take accurate kinetic readings from these samples. With a little troubleshooting, we realised it was possible that ACE2 was interacting with the NTA surface non-specifically. The easy fix? We switched to a CM5 sensor chip onto which an anti-His antibody had been immobilised, which completely removed the non-specific binding we observed for the NTA chip.
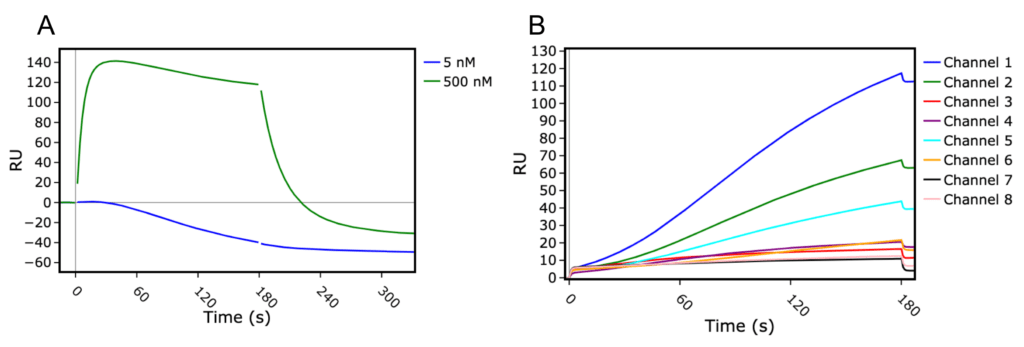 Figure 1 – Sensograms from test runs on the Biacore 8K. A) Sensograms from SARS-CoV spike (SARS1) with 5 nM (blue) or 500 nM (green) ACE2 used as analyte. These sensograms have been reference cell subtracted, causing a downward trend where there should be a flat plateau. B) Sensograms of reference cells in all 8 channels as ACE2 is being injected as the analyte. Increases in signal over the course of analyte injection in the reference cell is usually indicative of analyte non-specifically binding to the sensor surface.
Figure 1 – Sensograms from test runs on the Biacore 8K. A) Sensograms from SARS-CoV spike (SARS1) with 5 nM (blue) or 500 nM (green) ACE2 used as analyte. These sensograms have been reference cell subtracted, causing a downward trend where there should be a flat plateau. B) Sensograms of reference cells in all 8 channels as ACE2 is being injected as the analyte. Increases in signal over the course of analyte injection in the reference cell is usually indicative of analyte non-specifically binding to the sensor surface.
Measuring the kinetics
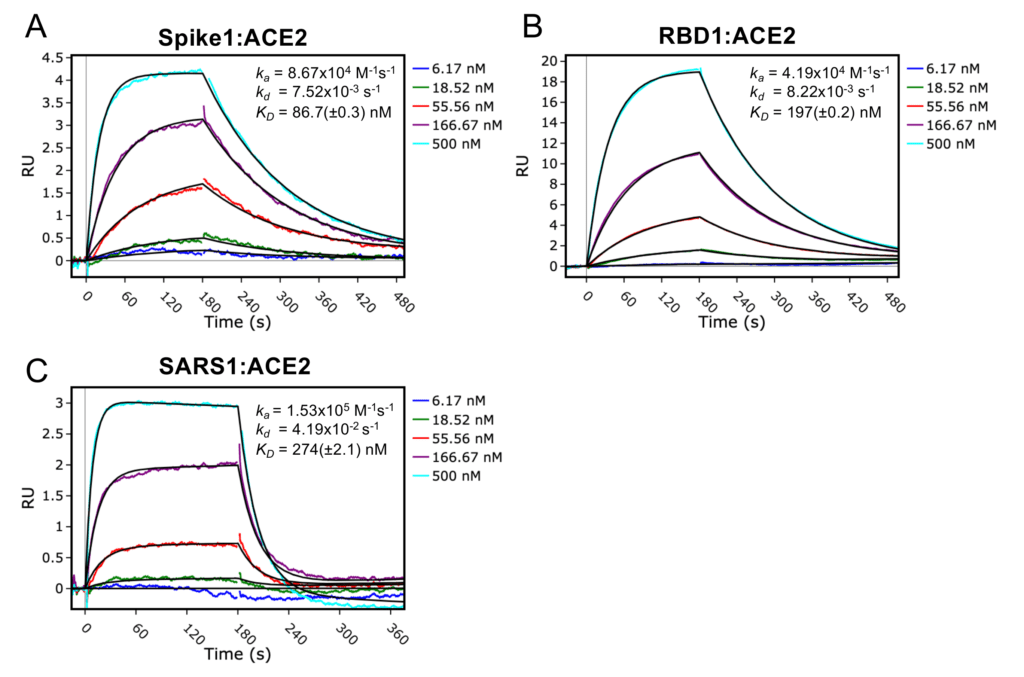
Once we had optimised the assay parameters, it was time to collect the kinetics and affinity measurements. In our assay, we had three S protein constructs to test: SARS-CoV-2 S protein (Spike1), SARS-CoV-2 S protein receptor binding domain (RBD1) and SARS-CoV S protein (SARS1). Each of these had six different concentrations of ACE2 flowed over them, and sensograms collected to measure the association and dissociation rates. Using the Biacore Insight Software, it was really quick and easy to analyse our results and fit a 1:1 binding model that agreed very closely with our raw data (Figure 2).
Wrap-up
With our current set-up, the affinities that we measured show a weaker interaction between SARS-CoV-2 homotrimeric spike and ACE2 than has been observed from the literature (Wrapp et al. 2020). Nevertheless, the trend we observe is consistent. The SARS-CoV-2 homotrimeric spike construct has the highest affinity to ACE2, while the RBD alone has a two-fold weaker interaction. In addition, we measured that the SARS-CoV homotrimeric spike has a 3-fold weaker affinity for ACE2 than the SARS-CoV-2 homotrimeric spike, one of the reasons postulated as to why SARS-CoV-2 is more infectious than SARS-CoV.
In conclusion, we were able to verify that our recombinantly expressed and purified coronavirus Spike proteins are folded and active, and can be used to develop or validate new assays. If you are interested in how Peak Proteins can help with your COVID-19 protein supply or production needs, please get in touch at info@peakproteins.com.


