Sherlock Holmes and the Case of the Missing Cysteines.
We had been asked by a client to express and purify a protein:Fab complex to support a Cryo-EM project. This was just to be a reproduction of a crystal structure that had been reported in the literature and so on the face of it looked to be fairly straightforward.
We received the target sequences and so began our campaign. However, fairly early on our suspicions began to be aroused. The protein target, itself a complex, was purifying well, with good yields and purity, but the Fab in which the heavy and light chains had been co-transfected into HEK293 cells, gave low yields of protein. With the majority of Fabs we produce, we see excellent yields and purity from the HEK293 system we use. Conspicuously, the Fab was running as a dimer on non-reducing SDS-PAGE.

Figure 1: Non-reducing PAGE of the original Fab expression (final sample, much concentrated)
We started our investigations to try and understand what was going on. Our first port of call was the PDB entry, where we wanted to check that the amino acid sequences we had been given for the Fab were correct. On first look they were identical, but on closer inspection one of our scientists noticed that neither the light chain nor the heavy chain contained the cysteines at the C-termini that are required to form the interchain disulphide bond [1]. Both sequences appeared to be terminated too early; the PDB sequence entries were incorrect.

Figure 2: Alignment of the C- terminal sequences given in the PDB for the Fab heavy and light chains, with the correct sequences that we subsequently cloned, which include the two cysteines required for the interchain disulphide bond.
We re-cloned both the heavy and light chain genes, with the offending cysteines included and shortly afterwards were back where we expected to be with excellent yields and purity of the intact Fab. This was able to stabilise the target protein complex and ultimately led to the successful generation of the Cryo-EM system.
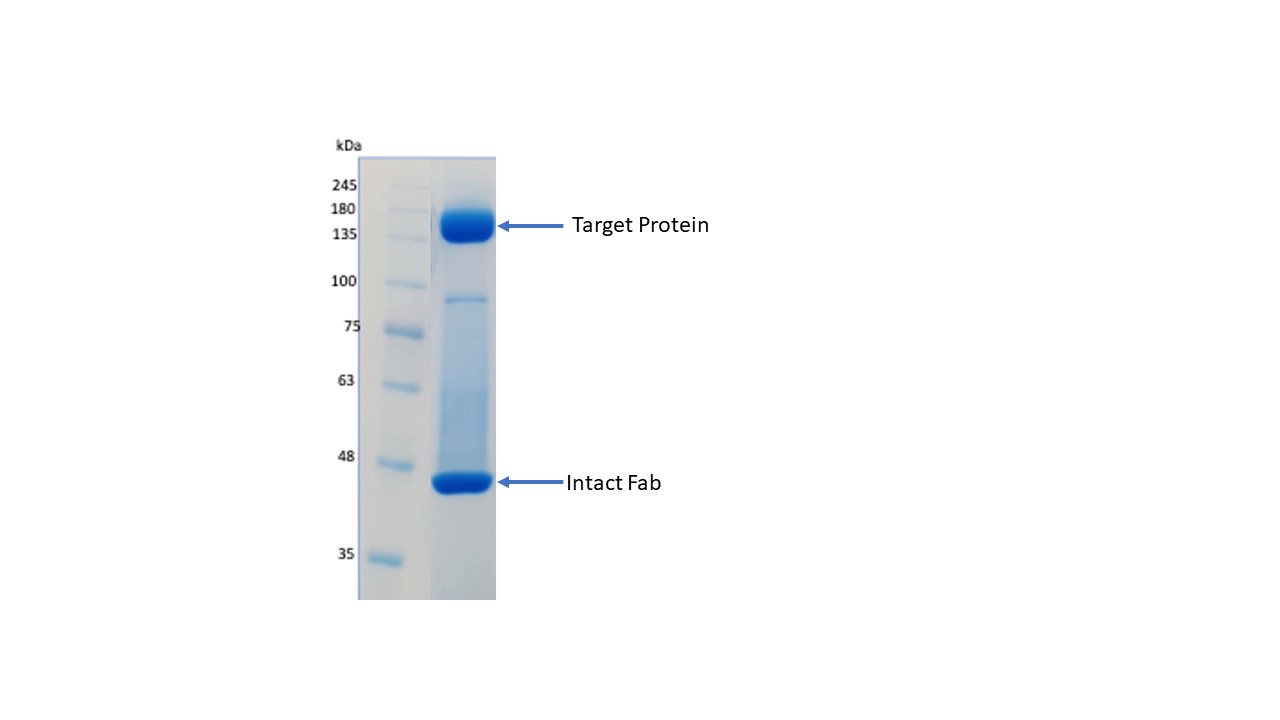
Figure 3: Non-reducing PAGE of the target protein complexed with the Fab, that was successfully used for Cryo-EM.
Reference
[1] H. Liu and K. May, “Disulfide bond structures of IgG molecules: Structural variations, chemical modifications and possible impacts to stability and biological function,” MAbs, vol. 4, no. 1, pp. 17–23, Jan. 2012, doi: 10.4161/MABS.4.1.18347.


