The Ligand That Can’t Decide…
Development of Selective Phosphatidylinositol 5‐Phosphate 4‐Kinase γ Inhibitors with a Non-ATP-competitive, Allosteric Binding Mode
Once Upon a Time
The vast majority of the structures we solve at Peak Proteins are strictly for the eyes of our clients only. We were however delighted that Steve Andrews and his team at the ALBORADA Drug Discovery Institute (ADDI) in Cambridge wanted to deposit structures, which we had determined as part of a joint project, into the public domain.
Once upon a time there was a protein target, Phosphatidylinositol 5‐Phosphate 4‐Kinase γ (PI5P4Kγ), which was one of a large, heterogeneous group of kinase and phosphatase enzymes responsible for the interconversion of different phosphoinositide lipids present in cells, thus involved in many aspects of cell physiology As well as being of interest to Steve and his group, as a target for neurodegeneration research, they are also attractive therapeutic targets in diseases such as cancer and immunological disorders.
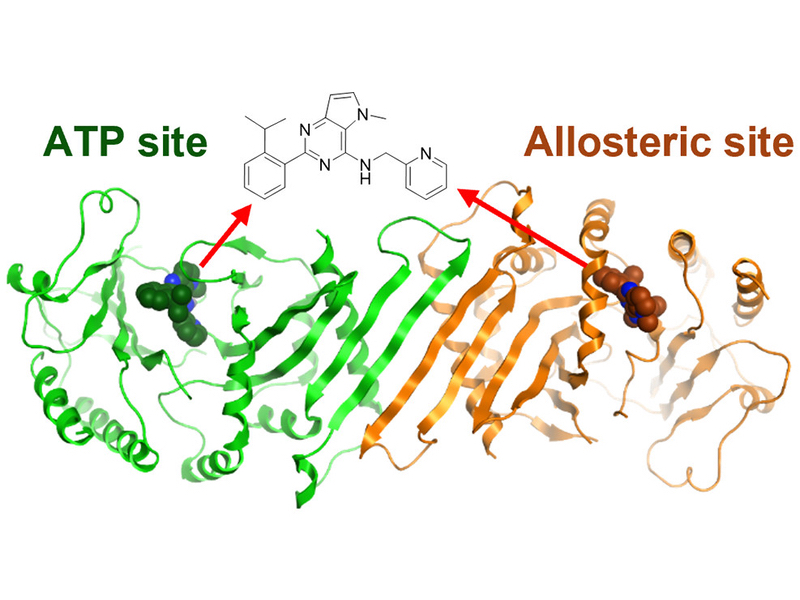
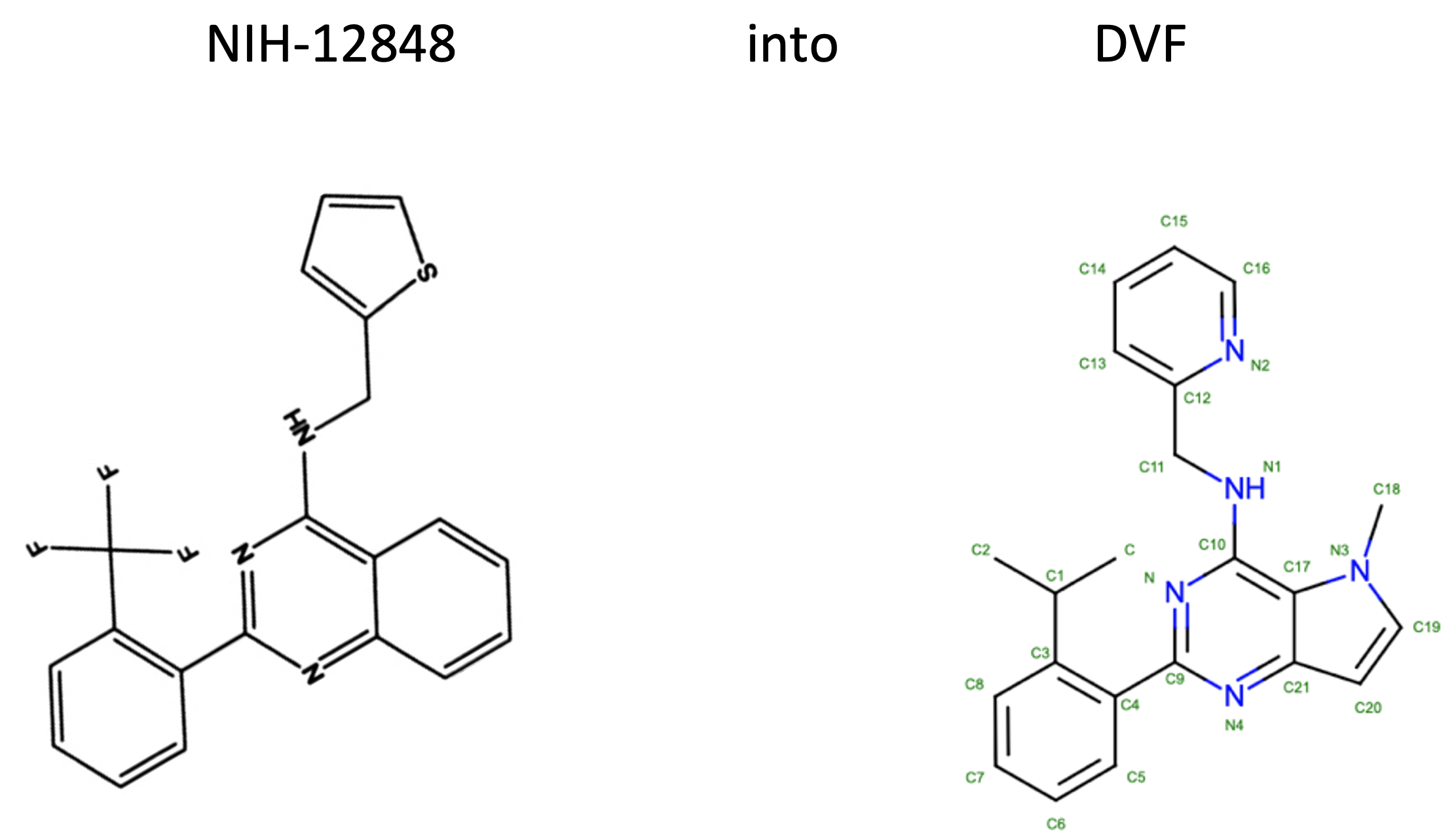
The joint publication describes how the scientists at the ADDI greatly improved the physicochemical properties of NIH-12848, a selective PI5P4Kγ inhibitor (ie. not inhibiting PI5P4Kα or PI5P4Kβ). At Peak Proteins we then solved the X-ray structure of the resultant ligand bound to PI5P4Kγ; the first X-ray structure of PI5P4Kγ complexed with an inhibitor. Not only did this structure confirm that the ADDI’s inhibitor was bound but showed that it did so in an allosteric manor. This allosteric binding mode is particularly interesting as it involves a putative lipid interaction site which is 18 Å from the ATP pocket.
So how did we all do this?
Structural backdrop
Structural information for each of the PI5P4K isoforms (a, b and γ) was already publicly available in the Protein Data Bank. However, none of these structures had drug-like ligands bound. The only crystal structure of PI5P4Kγ, at that time, (PDB: 2GK9 determined at the Structural Genomics Consortium – SGC) was of the apo protein solved at a resolution of 2.8Å. PI5P4Kβ on the other hand had structural information available where it was seen to be bound to a range of nucleotides (AMP, AMPPNP, GMP, GMPPNP). These allowed the ATP binding pocket to be modelled within the apo structure for PI5P4Kγ.
At Peak Proteins
Previous experience with this crystal system had shown that crystals diffracting to 2.8Å were in the difficult to produce reliably. Also there were several sizable stretches of the protein chain that were not resolved in the published structures, probably owing to protein disorder. We therefore set about designing modified constructs, removing potentially disordered regions, with the aim of improving the resolution and reproducibility.
Regions targeted were unresolved stretches of between residues 290-366 (the so-called “insert” region), the “activation loop” from residues 377-403 as well as a number of N-term truncations. Settling on a list of 5 constructs (including the SGC precedent), we obtained good expression for all these in E. coli. All constructs ran as dimers on size exclusion traces and, were all put into crystallisation trials.
Broad crystallisation screening, along with optimisation of SGC conditions, produced sub 3Å diffraction for the best 2 loop deletion constructs (Interestingly these were the longest and shortest deletions). These crystals were obtained using convenient sitting drop set-ups, in addition to the more labour intensive hydrogel method reported by the SGC. Crystallisation conditions were chemically similar to those for the original SGC structure but with the use of crystal seeding techniques we were routinely able to obtain apo crystals that diffracted to around 2.4Å resolution.
Meanwhile down in the south…
The ADDI the team were working on development from NIH-12848. With an in vitro IC50 of approximately 1 μM against PI5P4Kγ and being a lipophilic molecule (CLogP 6.5) with poor solubility (<5 μM) there was plenty of scope for improvement. Attempts to computationally dock NIH-12848 into the postulated ATP pocket in a convincing binding pose had been unsuccessful so their optimisation was initiated using classical medicinal chemistry approaches.
The SAR (structure activity relationship) was determined for 27 different small substituents introducing changes to the trifluoromethylphenyl or thiophene groups and on balance, from this initial set of thiophene replacements, a pyridine ring was found to be preferred.
More variations of the quinazoline core were undertaken, including truncating it, deleting lipophilic atoms or replacing them with heteroatoms. Overall a pyrrolopyrimidine core was selected and the molecule destined for crystallography experiments was chosen (shown below, DVF was the ligand ID assigned by the PDB on deposition of the structure).
So back to the North…
As soon as the carefully crafted ligands arrived at Peak Proteins, we set to work and were successfully able to obtained co-crystals, using apo protein crystals as seeds. Then after a trip to the Diamond Light Source in Oxford to collect X-ray diffraction data we obtained liganded structures in the 2.0 – 2.5 Å range.
The rest of the story is best told in pictures…
… and they all lived happily ever after, with the crystal structure showing the inhibitor bound at the two different binding sites within one protein dimer. PDB:7QIE PI5P4Kγ bound to DVF at 2.4 Å resolution.
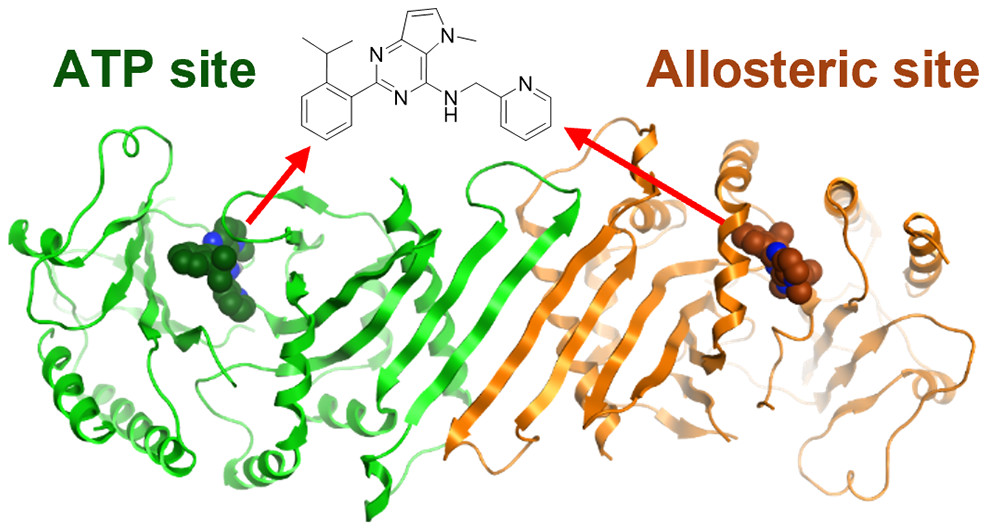
Dimer of PI5P4Kγ chains A (orange) and B (green) with DVF bound
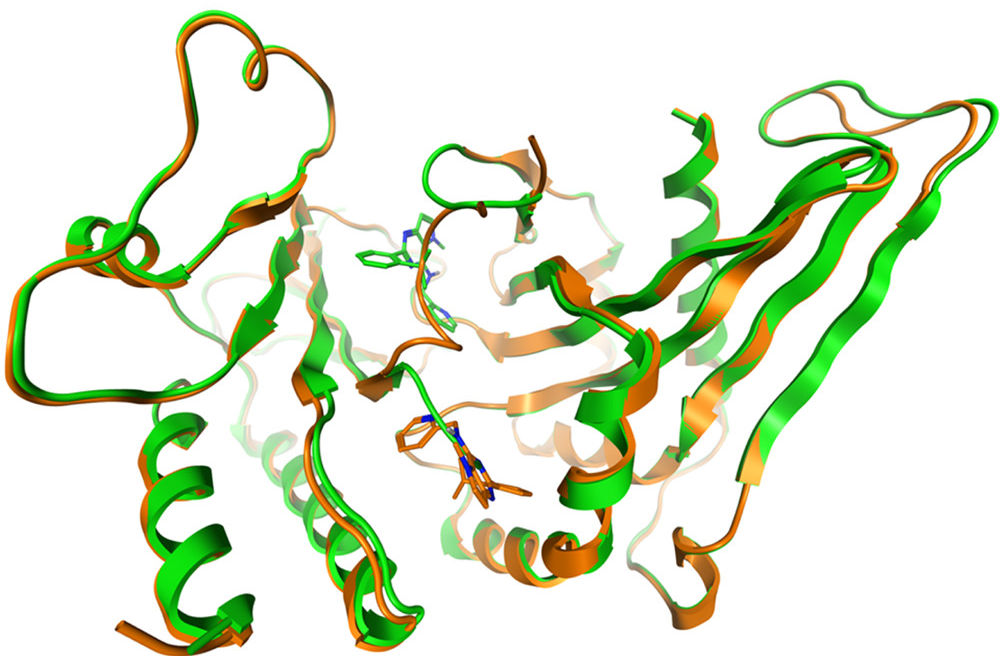
Two binding sites for DVF: chain A (orange, DVF in allosteric binding pocket) and chain B (green, DVF in ATP site) superposed with DVF in the stick. The binding sites are mutually exclusive