The Journey To Solve a Challenging Protein Purification
In this case study, Toby Allen, one of our protein scientists leads us through the various steps we took to resolve a particularly tricky protein production challenge.
We worked on both a full length and truncated version of a DNA binding protein and employed a variety of tags and purification methods to finally be able to generate the target. This is a really good example of what we do best at Peak Proteins; treating each protein as a unique challenge, responding to the data we obtain and designing our next experiments accordingly.
THE CHALLENGE
At Peak Proteins, we have been working to develop a robust expression and purification protocol for this DNA binding protein which is required to support a wider client project within Sygnature Discovery. It has proved to be an incredibly challenging protein to work with.
Experiment 1: The initial attempt
Our initial attempt was based on a relatively sparse literature precedent and ultimately resulted in low yields and instability.
There was just one paper available that reported purification of both a FLAG tagged full length (FL) and a His tagged truncated version of the protein that were expressed in insect cells. An interesting quirk in the protocol was the inclusion of a LiCl wash to reduce the level of impurities. An SDS-PAGE gel of the final sample showed the FL construct at a purity of around 60%, so we decided to pursue this approach first.
In our hands we obtained relatively pure protein from the FLAG affinity column, (Figure 1) with one main lower molecular weight contaminant. But we observed protein precipitation in the final sample, that explained the low yield (<0.1mg purified protein / Litre of culture) that we obtained.
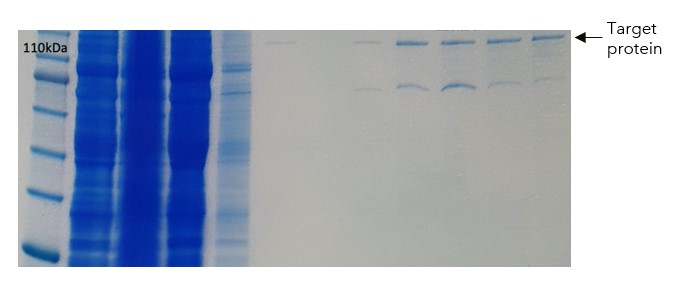
Figure 1: PAGE showing results from initial expression and purification on FLAG resin
Experiment 2: Removing the LiCl
We hypothesised that the LiCl wash was resulting in protein loss and therefore, a run was performed without this step. This did increase our recovered yields (no precipitation was observed), but unfortunately led to significantly more contamination. (Figure 2).
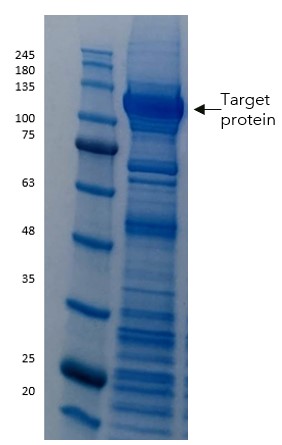
Figure 2: FLAG elution pool where LiCl wash step had been removed
Experiment 3: Switching Li for K
In order to try and reduce the contamination and maintain stability of the protein, we attempted washing with KCl instead of LiCl. This resulted in a significant increase in yield (now 0.7mg protein per L culture) compared to the initial prep and the purity was much improved.
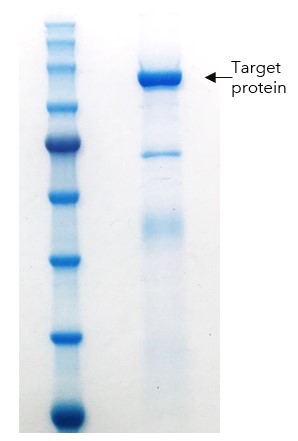
Figure 3: FLAG elution pool with KCl used in the wash buffers instead of LiCl
This represented a significant achievement, but issues seen in the original prep remained. The material was still too unstable to concentrate post FLAG-affinity, and the lower molecular weight contaminant was still present. Additionally, the material was too dilute to process via size exclusion chromatography that ideally we wanted to include as a final polishing step.
Experiment 4: Switching constructs
We had already cloned the other 6His tagged, truncated construct from the paper and at this point decided to try this version. We also added an AVI tag to allow biotinylation of the target that was required for immobilisation in surface plasmon resonance (SPR) experiments.
This construct was also expressed in insect cells but unfortunately subsequent eluted fractions from the (Immobilised metal affinity chromatography) IMAC column were very impure (Figure 4)
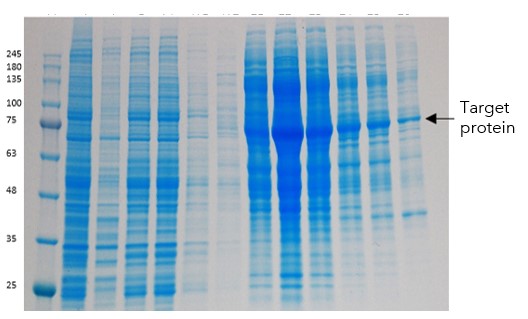
Figure 4: PAGE showing Load, wash and then elution fractions from IMAC chromatography for His tagged truncated version of the protein
Again, even though from the initial data it looked like we had achieved good yields, extensive precipitation was observed on elution from the IMAC column, resulting in the total loss of the target protein.
Experiment 5: Switching tags
Our next suggestion to try and improve the purity was to use a twin-Strep tag (TST) instead of the His tag. We also included a TEV site to allow removal of the tag for downstream purposes. Streptactin resin was then used to purify the protein.
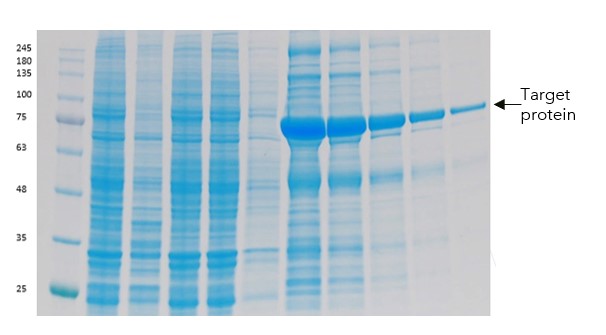
Figure 5: PAGE showing Load, wash and then elution fractions from Streptactin chromatography for the TST truncated version of the protein
The results were not only increased purity (from the higher selectivity of the twin-Strep tag), but also good stability and yields at this stage.
The Twin-Strep tag was removed by TEV cleavage, and the protein further processed by concentration and SEC. No stability issues or degradation were observed and an accurate A280 could be obtained (Figure 6).
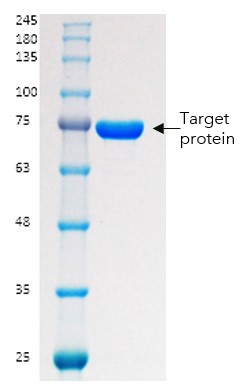
Figure 6: PAGE of final purified truncated target protein sample
Purity was >95% and a yield of 0.7mg protein per L of culture was obtained. Biotinylation was confirmed by streptavidin gel shift assay (Figure 7).
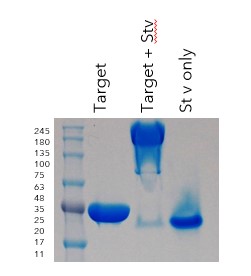
Figure 7: Streptavidin (Stv) has bound to the biotinylated AVI tag on the target, shifting the band to >250kDa.
THE IMPACT
We were able generate the DNA binding protein (including successful cellular biotinylation). Over the course of the work, we were able to improve the yield, purity, and stability of the target. This is where we believe we stand out at Peak Protein – the ability to design strategies, react and respond to data, and ultimately deliver. Contact us info@peakproteins.com to learn more about what we offer.


