Using Mass Spectrometry to Rapidly Facilitate Covalent Drug Discovery
The Name’s Bond… Covalent Bond.
Covalent compounds have historically accounted for a significant number of successful drugs, but interest waned due to concerns about poor selectivity that is often identified in early assay screening. One of the key hurdles in a covalent drug campaign is being able to identify successful covalent modification at a pace which can be used to iteratively inform the chemistry.
In this case study, one of our Senior Protein Scientists, Alex Brown, shows how intact mass spectrometry can be used to rapidly identify successful covalent binders, but beyond this – also to infer specificity characteristics of each compound.
A reliable supply of well-behaved, high-quality protein is the starting point. For this reason, protein was expressed and purified in-house for this project, which gave repeatable results using protein from different batches.
Rapid decision making
Before more detailed information is required, answering the question – “does this compound bind covalently to the desired protein” is the first step. For this, a suitable starting concentration and ratio of compound: target protein is determined based on the desired parameters for the individual project. A large number of compounds can be tested at this stage, quickly enough to inform chemistry decisions, with only the most promising going forward into more detailed trials.
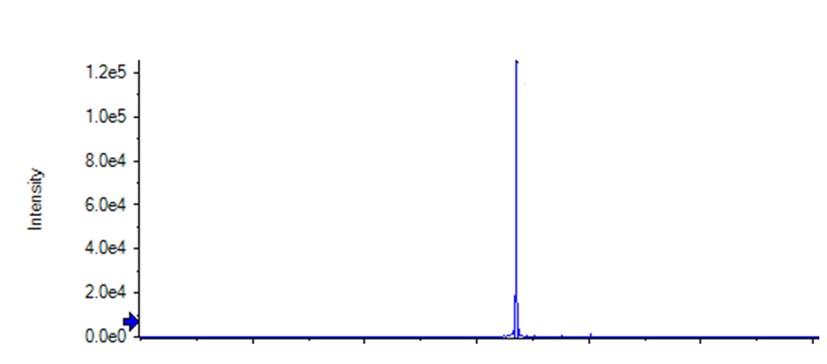
Figure 1: Intact mass spectrometry – target protein only sample showing a single peak.
Figure two below shows the mass spectrometry of the target protein incubated with two different compounds from the chemistry pipeline. Intact mass spectrometry will not show any change in mass from non-covalent interactions. The spectra of both compounds include a peak corresponding to a mass of bound compound (protein + compound) – 18 Da (leaving group = H2O) signifying successful covalent modification. However, there are evident differences between the spectra.
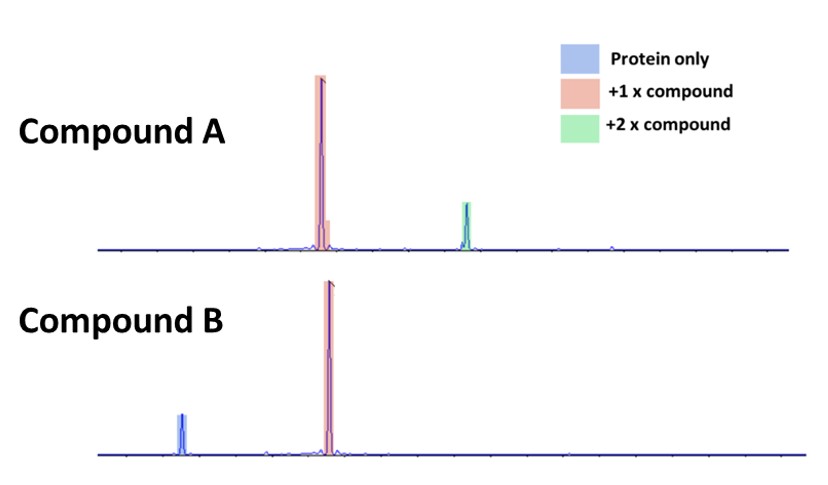
Figure 2: Intact mass spectrometry of the target protein incubated with two different compounds
The activity/specificity relationship
Compound A shows a complete conversion of the protein to bound protein – there is no protein-only signal. However, it also has a peak which corresponds to protein bound to two compounds.
Compound B by comparison still has a signal for unbound protein, along with a larger peak for single compound bound to protein.
These two compounds demonstrate the activity/specificity relationship that we can investigate with intact mass spectrometry of covalent compounds, and the information that can be gained by titrating compounds of most interest. We can tell from these spectra that the compounds are behaving differently but not which would make the better drug candidate.
Compound A shows better binding, but the +2 signal shows that the compound will bind to two different locations on the protein, indicating non-specificity – potentially very damaging for a drug prospect. Without a protein only signal, it is possible that the binding is good enough that the concentration of the compound can be reduced substantially – reducing any excess binding to a negligible point and still completely binding the protein.
Conversely, the lack of a +2 signal in the compound B sample does not necessarily mean the compound is more specific, increasing the concentration to completely bind the protein might incur the +2 or higher states. Determining this activity/specificity relationship can rapidly identify the most suitable compounds to develop in a project, saving costs associated with chemistry of non-suitable targets, and allowing resources to be funnelled towards the targets with the best potential.
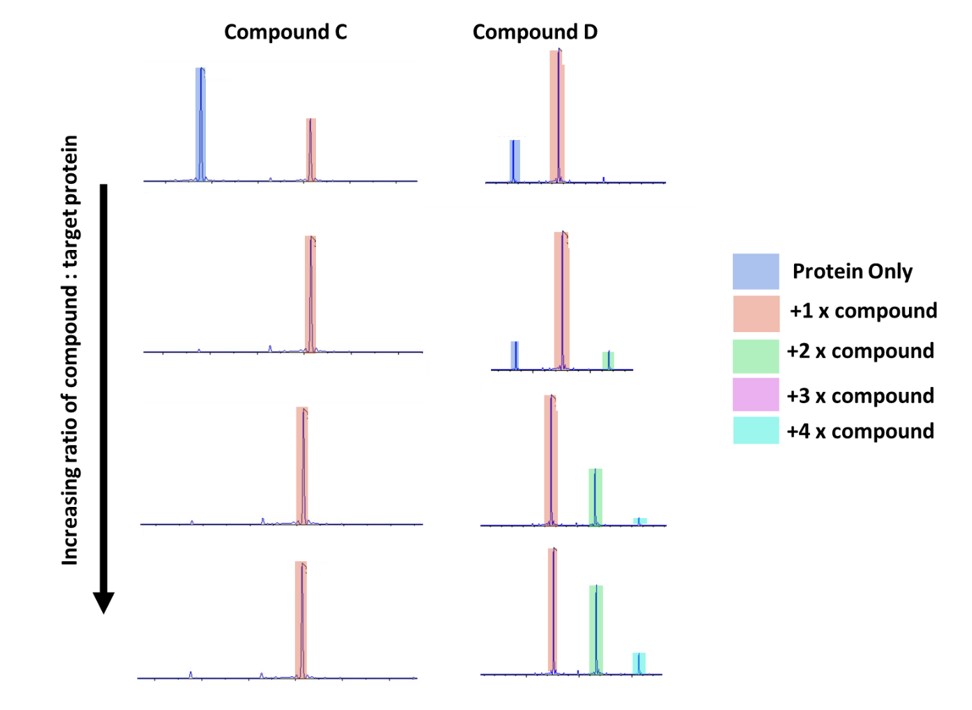
Figure 3: Intact mass of two different compounds compared.
At the lowest ratio of compound: protein these look similar (Fig. 3), but by performing the titration of increasing concentration it is easy to see that compound C looks to have excellent specificity – even with a high excess of compound there is only binding to one location on the protein. This can be identified as a good target going forward.
Which binding site?
The previous experiments demonstrate preference for one binding site over others but at this point we cannot be sure that the +1 is representative of binding to the desired site.
For this project there were three techniques used to prove that this was the case, cross testing against human serum albumin, competition assay using a well-characterised non-covalent inhibitor and finally a mutation of the desired binding site. Each of these methods required different levels of resourcing and effort and are therefore suitable for different points of the drug discovery journey.
Human serum albumin
In this project the protein-compound interaction was in human blood, making human serum albumin one of the most prevalent non-target proteins. After brief, initial testing to quantify a suitable concentration for mass spectrometry, this was performed in parallel with the rapid testing of all compounds to provide additional information i.e. do the additional bound states of the target protein translate to binding to non-target proteins as well.
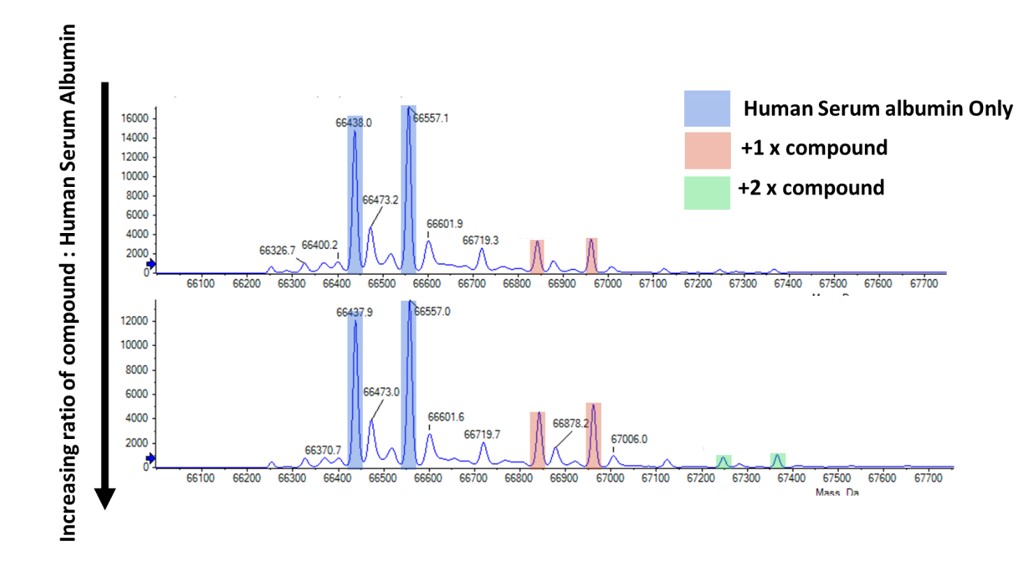
Figure 4: Intact mass spec example of commercial HSA and compound binding. Note that the commercially produced HSA had two dominant species – cysteinylated and non-cysteinylated resulting in the two peaks.
The next method of analysing the covalent binding was to use a known non-covalent binder to out compete the covalent one. This non-covalent may be part of the current drug campaign or an available commercial tool drug. Logically, if this non-covalent compound is added while the covalent compound remains at the same concentration and the binding seen on the intact mass is reduced, there is competition for the same binding site. If the binding site targeted by the non-covalent is known to be the desired site, then this results in more data to suggest the covalent is also targeting the desired site.
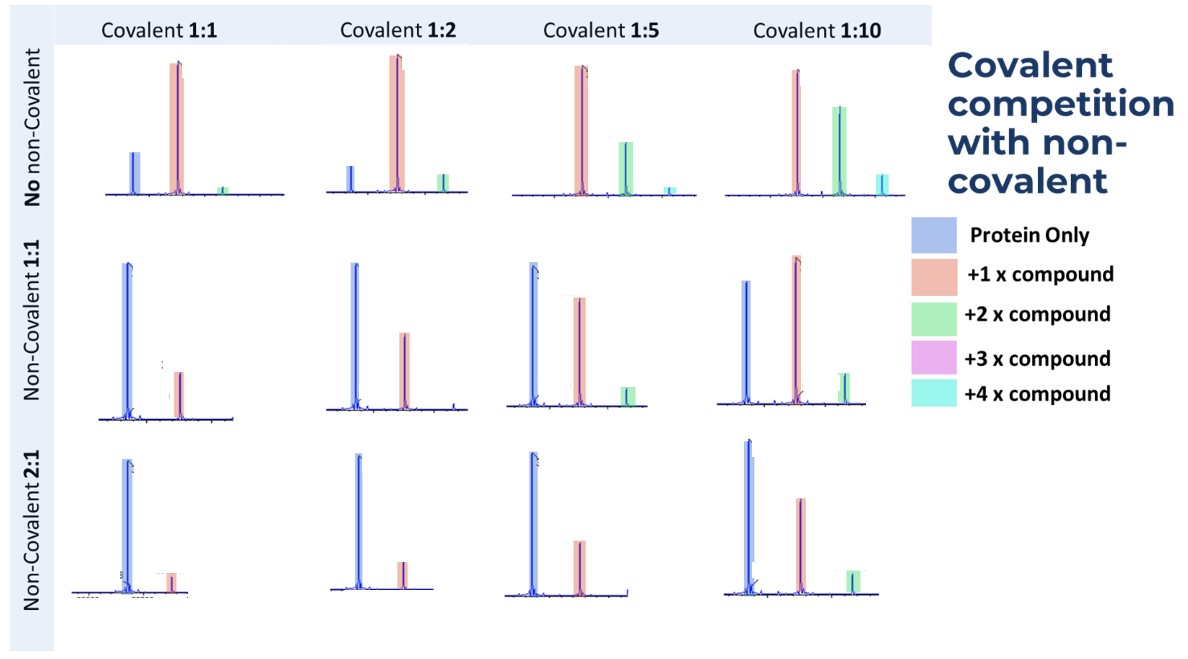
Figure 5: Results of a titration with the target protein and the target compound.
In this case the covalent compound appears to have some non-specific binding (+2, +3, +4 states), but is also being outcompeted by the non-covalent binder (data shown for when no non-covalent binder is present, then 1:1 and 2:1 ratios)
Confirmation of binding to target site by testing Vs a mutant version
The most robust system to prove that the binding is occurring at the target site, or particularly that the covalent modification is to a target residue, is by creating a deletion mutant of that residue (Fig. 6). At Sygnature we are well positioned to undertake complimentary approaches like this, particularly if the target protein is also being expressed by ourselves as we will have a well-defined purification protocol – again saving time and money on optimisation.
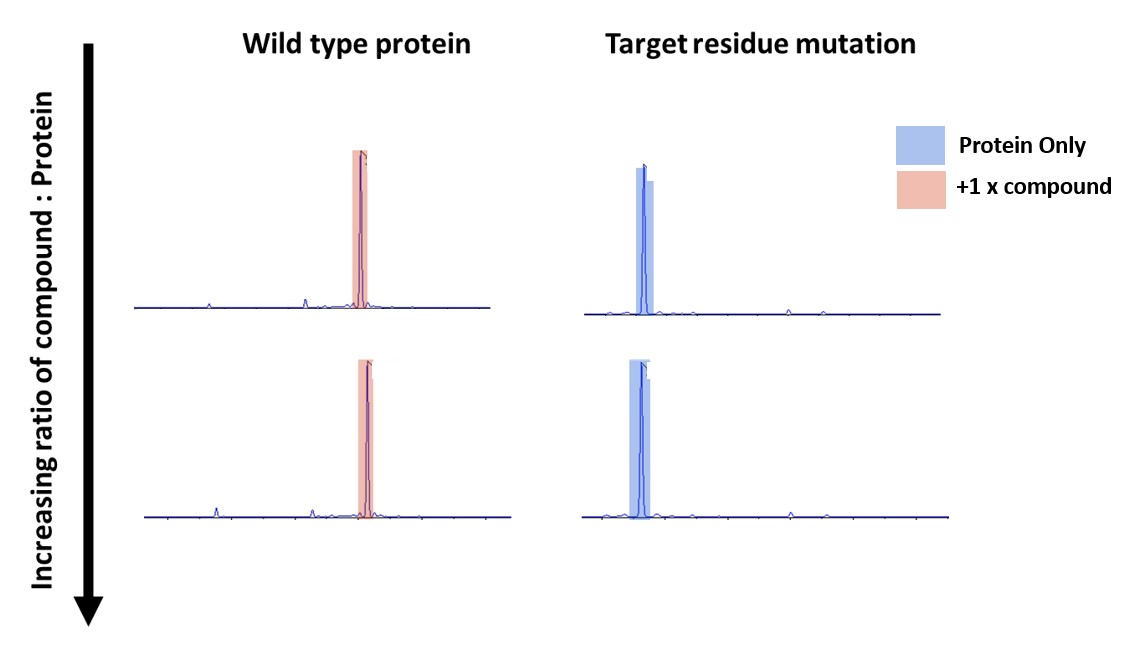
Figure 6: Intact mass of Wild type and Mutant protein that have been incubated with covalent compound
The results above shows that as before the wild type protein is binding 1 compound molecule and that the mutated protein does not bind to the compound at all. Thereby demonstrating the binding is occurring at the target site.
One concern is that this mutation will affect the structure of the protein, which in this project was alleviated because the protein was being used in a very well characterised crystallography structural system. Therefore, we crystallised the mutant in the known conditions, and it was relatively trivial to obtain a high-resolution structure, align that with previous structures of the wild-type protein and show that there were no structural changes between constructs. Furthermore, in this case the resolution was high enough to actually see that the desired residue has been mutated. This is another example of how a ‘one-stop shop’ for supporting drug design allows for more robust experiments by combining expertise.
Summary
Despite early reservations about covalent compounds as drugs, due to concerns about specificity, recently interest has increased. Targeting non-conserved amino acids, to increase specificity, has become more popular. Here we show at Sygnature Discovery how we can use intact Mass spectroscopy to determine covalent binding of compounds, the number of binding sites and the location of binding. These experiments assist with hit generation and the hit to lead process of drug discovery. Lastly, we are able to test the compound binding to non-target proteins, helping to develop a specific and well characterised drug.


