Protein NMR and its role in Drug Discovery
We are fortunate at Sygnature Discovery to be one of the few CROs that offer all 3 of the main protein structural biology techniques: X-ray crystallography, cryo-EM and NMR . In this blog we give an overview of how we use NMR to support drug discovery programmes for our clients. This includes the Hit Identification phase where we support the “Hit Synergy” platform we provide at Sygnature.
Understanding how small molecules interact with proteins is central to drug discovery and Nuclear Magnetic Resonance (NMR) offers a uniquely sensitive approach to probing these interactions. Whether you’re screening fragments, validating hits, or exploring binding kinetics, NMR provides insights that complement other structural techniques and help guide decision-making throughout the discovery pipeline.
What is NMR?
Nuclear Magnetic Resonance (NMR) is a technique that exploits the magnetic properties of certain atomic nuclei — most commonly hydrogen (1H), carbon (13C), nitrogen (15N) ), and phosphorus (31P) — to gain detailed information about molecular structure and dynamics.
Unlike crystallography, protein NMR allows the investigation of proteins in solution, thereby more closely mimicking physiological conditions. Furthermore, it can cope with folded, intrinsically disordered and highly dynamic proteins. This makes it ideal for probing dynamic processes such as conformational changes, binding kinetics, and allosteric modulation. The technique does of course have its limitations. (see our structure technique comparison blog for more details). In general, the protein has to be produced in E. coli, cell free or insect cell systems to allow specific labelling. However, for ligand observed (LO)-NMR the protein can be unlabelled and therefore produced in any expression host cell line.
NMR in Drug Discovery
Protein NMR is a versatile and sensitive technique that plays a valuable role in small molecule drug discovery. It enables researchers to observe molecular interactions in solution, offering insights into binding events, kinetics, and conformational changes, often in ways that other structural methods cannot. Below are some of the key ways we apply NMR in our drug discovery workflows:
- Protein:Ligand interactions
- Ligand observed: Especially useful for orthogonal confirmation of binding and drug design
- Protein observed: Elucidating the actual binding site
- Investigating dynamic process upon compound binding
- Binding kinetics in real time, even for weak binders.
- Monitoring conformational changes
- Monitoring allosteric modulation
- Competition binding
- 3D structure of proteins and protein complexes
- Protein:ligand binding site described. To inform medicinal chemistry in the design – make – test cycle.
- Identification and characterization of cryptic and allosteric pockets not visible by other techniques.
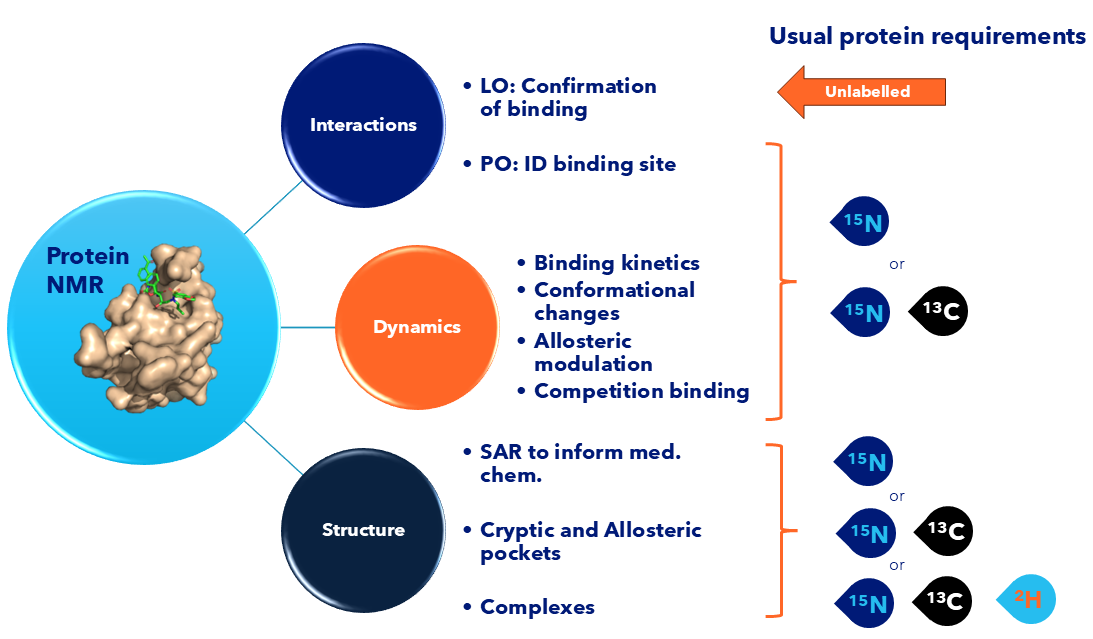
Figure 1: Overview of Protein NMR applications in drug discovery. Specific labelling required depends on the size of the target protein and the particular experiment
Interactions with ligands
Protein NMR is especially valuable as a follow up in fragment-based drug discovery (FBDD), where small molecular fragments are initially screened for binding to target proteins. We then use ligand observed LO- NMR to confirm binding of the fragments using unlabelled protein and follow up with protein observed PO-NMR (requiring uniform 15N labelled protein) aiming to identify the site of binding at the amino acid level. This can be a critical part of the hit triage process and allows identification of ‘true binders’ which can then be used to validate other elements of the screening cascade (e.g., SPR). In our experience this has enabled many projects, especially those that have had no tool compounds.
NMR in solution enables the identification of low-affinity binders (kD = µM-mM), as well as the characterisation of their interaction sites. In addition, NMR allows better understanding of whether hits target the same site or different sites through competition studies. These data can then guide the rational design of more potent compounds.
LO-NMR NMR methodologies allow the identification of hits in a reasonable time frame of few hours (0.5-4 h) depending on ligand solubility. A typical initial experiment will just look to define if the ligand is a simple “binder” or “non-binder”.
Binding kinetics
Studying protein:ligand binding kinetics using NMR provides several advantages over other static techniques. We often recommend using NMR as an orthogonal method during a drug development program.
Real-Time Monitoring: Protein NMR can monitor and characterise the binding modes of the ligand in real time. This allows direct measurement of not just equilibrium affinity constants (KD), but also association (on-rate, kon) and dissociation (off-rate, koff) rates. This is critical for understanding how long a drug remains bound to its target — a key determinant of efficacy. Data obtained can therefore allow the ranking of hits to help inform further chemistry.
Physiologically Relevant Conditions: Furthermore, NMR is performed in solution, with the aim being to mimic as closely as possible the physiological environment of the target protein. This makes the observed kinetics more representative of in vivo conditions and is a key advantage over techniques such as SPR where the protein is immobilised on a chip, where sometimes the binding sites are spatial occluded from the solvent as result of the immobilisation strategy. The protein can also be tag free, avoiding any issues where fragments or ligands are suspected to bind to tags or linkers.
Protein Structure
To determine the structure of a protein target will require the production of 15N, 13C labelled (and potentially partially 2H) protein.
Structure-activity relationship (SAR) studies: When it is possible to determine the NMR structure of a protein target with ligands bound, it provides excellent data to support medicinal chemistry structure-activity relationship (SAR) studies . Modifications made to ligands are evaluated to see how they affect binding, thereby helping to drive the design – make – test (DMT) cycle. A lot of information can be gained from just resonance assignment, particularly if there are known structures or structures of homologous proteins. This allows identification of binding site and can guide in-silico modelling studies.
Allosteric and Multi-Site Binding: NMR’s atomic resolution allows detection of subtle shifts in atomic environments. This can reveal complex binding mechanisms such as allosteric interactions, conformational selection, or induced fit. These types of insights are simply not available using other techniques.
Cryptic Pockets: We have also had success using NMR to identify a novel cryptic pocket in a cytokine. Watch this space for more information in the near future.
Pros and Cons of NMR
| Pros | Cons |
|---|---|
|
|
Protein NMR continues to be a vital tool in the drug discovery toolkit — especially when flexibility, sensitivity, and solution-phase insights are needed. At Sygnature Discovery, we combine deep technical expertise with a tailored approach to ensure NMR delivers meaningful data for each unique project. If you’re working with challenging targets or need complementary structural insights, our team is ready to help. info@peakproteins.com