Producing Homotrimeric Proteins
Many proteins exist and function as multimers in their native state. However, the mechanism by which they achieve this can prove to be an issue when trying to express and purify the protein recombinantly.
Two recent projects we have been involved with, required solutions to this in order to produce homotrimeric proteins.
Foldon fusion
The first was the SARS-CoV-2 spike protein which is active as a homotrimer. To produce the protein recombinantly, teams have replaced the transmembrane region with a bacteriophage T4 fibritin trimerization motif [1] (aka “Foldon b-propeller”) [2] [3] [4] [5] along with other stabilising mutations. We successfully reproduced these methods in the Peak Proteins’ laboratories to generate homotrimeric spike in HEK293 cells. This purified with good yields, to produce pure, active spike as demonstrated by its binding kinetics to the ACE2 receptor in Biacore surface plasmon resonance (SPR) experiments.
View some of the work done by Peak Proteins:
Measuring coronavirus Spike:ACE2 binding affinity and kinetics using the Biacore 8K (Poster)
Measuring Coronavirus Spike:ACE2 Binding Affinity and Kinetics Using the Biacore 8K (Blog)
Measuring Coronavirus Spike:ACE2 Binding Affinity and Kinetics (Talk)
Furthermore, the spike protein we produced was used by Phelan et al 2021 [6] in their study “Dynamic Assay for Profiling Anti-SARS-CoV-2 Antibodies and Their ACE2/Spike RBD Neutralization Capacity”.
3HB fusion
For the second project, we were asked to produce a different protein as a homotrimer. This particular target relies on interactions with other proteins to trimerize in vivo, but for this project the client wanted the target protein on its own, in trimeric form. There are several trimerization motifs that have been used in the literature and we chose to compare two in parallel. The T4 foldon motif, as it had worked so well for spike, and the engineered 3 helix bundle (3HB) developed by Fletcher et al [7]. We expressed both constructs (with 6His tags and either Foldon or 3HB motifs) in E.coli and purified using metal affinity and size exclusion chromatography. Both expressed well with yields between 35 to 55 mg per litre culture. Interestingly, a significant proportion of the construct with the foldon appeared as aggregated peak running at the void volume of a calibrated size exclusion column. Whereas the main peak for the construct with 3HB ran with a calculated apparent mass of 54kDa which is expected for the desired homotrimer (Figures 1a, 1b and 1c). When pooled the homotrimer was found to be stable to both concentration and freeze thaw.
While both constructs produced homotrimeric protein, it was certainly worth comparing different trimerization motifs to find the one that gave the best yield, 3HB in this case.
Fletcher et al [7] described other variations of coiled-coil peptide oligomers to produce dimeric, and tetrameric proteins as well as the trimeric version used in this study.
Figure 1a: Overlay of size exclusion chromatography column traces. Blue: construct with 3HB motif, Green: construct with foldon motif
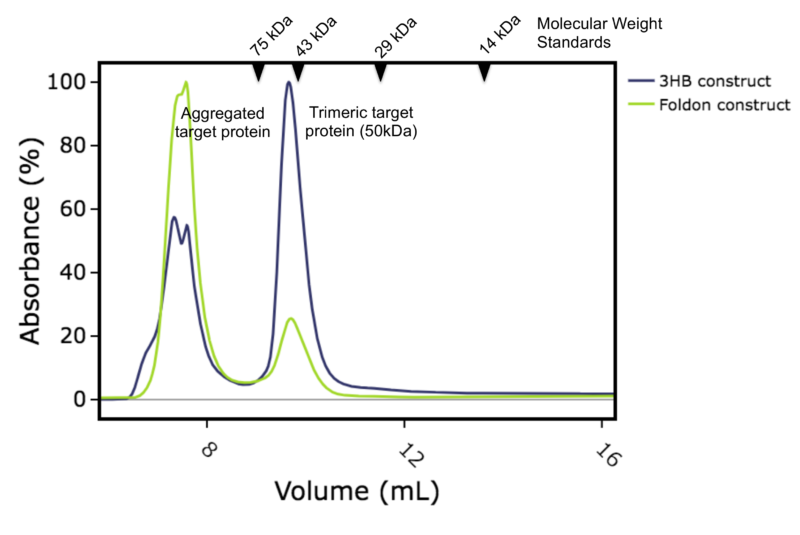
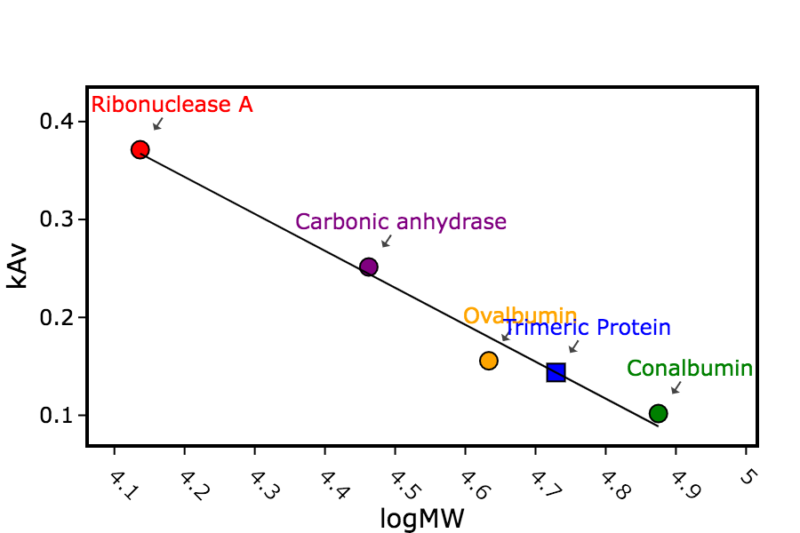
Figure 1b: Overlay of size exclusion chromatography column traces. Blue: calibration standards with molecular weights in kDa shown, Orange: construct with 3HB motif, Green: construct with foldon motif.
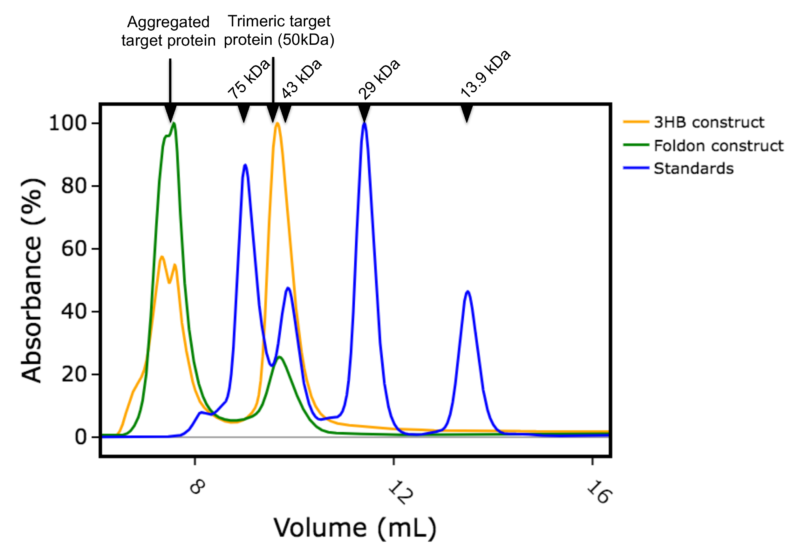
Figure 1c: Calculation of the apparent mass of the size exclusion peak for the target homotrimeric protein (with either foldon or 3HB trimerization domain)