Harnessing the Power of the Metabolome – What Can Metabolomics Potentially Do for Us?
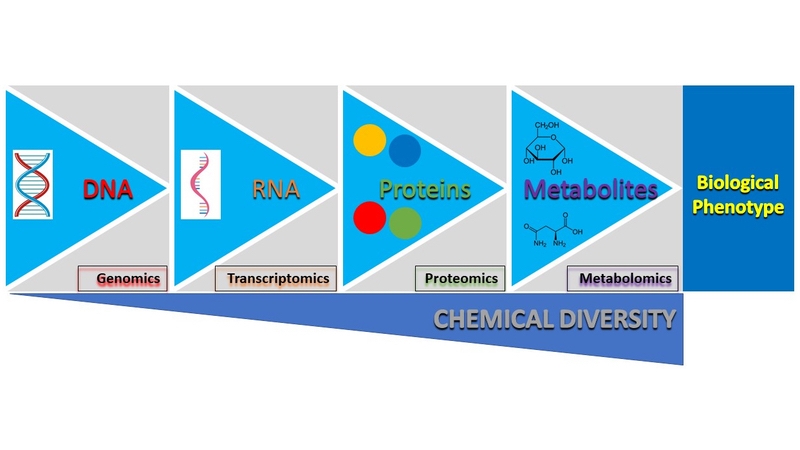
At Peak Proteins, we use mammalian, insect and bacterial cells grown in specialised culture media for the expression of recombinant proteins. A key focus for us is to ensure the expression systems we use are running optimally, in order to generate high yields of recombinant proteins. Given the complex nature of cells growing in a cultured environment, understanding what those optimal conditions are isn’t straightforward. Understandably, various systems biology tools are continually being developed and used to study and understand cell growth and/or protein production (Figure 1).
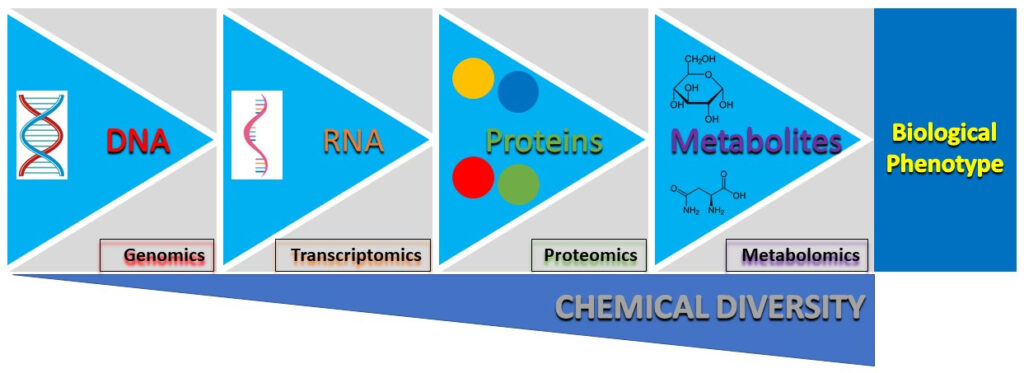
Figure 1. Central dogma of biology. A flow diagram showing the path from DNA to the biological phenotype. Highlighted at each stage is the corresponding systems biology tool used to study that particular ‘omics’.
The ability to analyse the spent media (extracellular metabolites) or cellular metabolites (intracellular) produced or used by cells during recombinant bioprocesses offers the ability to identify and replenish key nutrients that become limiting for cells during cell culture. These insights into the metabolome can offer a powerful indication of “cellular status” or “cellular health” which can provide information on molecular events that determine, regulate or limit cell specific functions such as cell growth and/or recombinant protein production.
Metabolomics, which is a relative new systems biology tool, can be defined as “the comprehensive profiling of all the biochemicals and metabolites in a biological sample. So, metabolomics is the study of cells through measuring their metabolite profiles or their so-called metabolic signatures. Each cellular metabolome contains hundreds of different metabolites which contains diverse chemical properties or species. Therefore, it’s not possible for a single analytical approach, as of yet, to be able to capture or take a snapshot of the entire metabolome in one go. Therefore, different, but complementary techniques are used to analyse the diverse chemical species of the metabolome. For both mammalian and insect cell metabolomics, the three most commonly used approaches are Nuclear Magnetic Resonance, Liquid Chromatography–Mass Spectrometry or Gas Chromatography–Mass Spectrometry. Each of these specific analytical techniques offers unique advantages and disadvantages over the others such as sensitivity and ability to specifically measure amounts of metabolites (Figure 2).
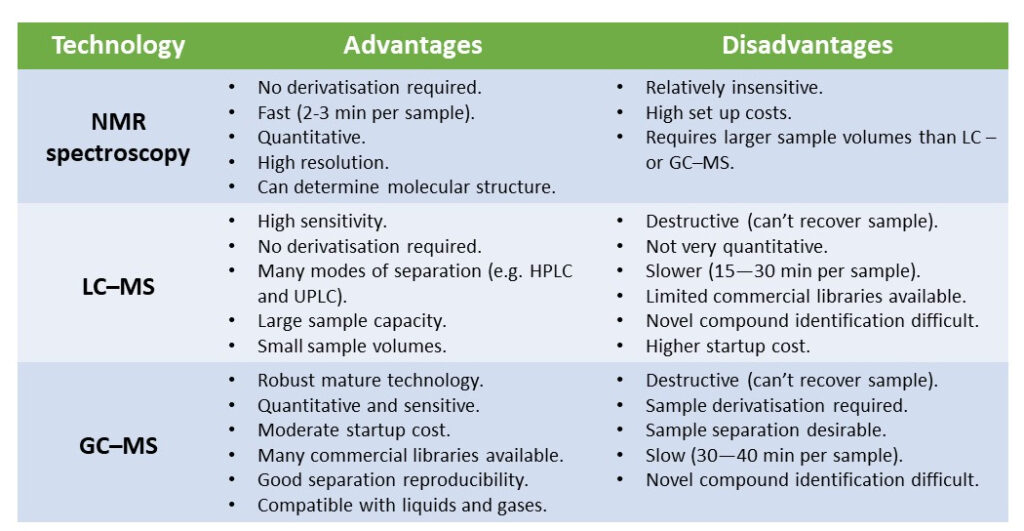
Figure 2. A comparison of analytical techniques used in metabolomics studies. Abbreviations: NMR (nuclear magnetic resonance); LC–MS (liquid chromatography–mass spectrometry); GC–MS (gas chromatography–mass spectrometry); HPLC (high-performance liquid chromatography); UPLC (ultra-performance liquid chromatography).
Mass spectrometry (MS)-based metabolomics methodologies have been used extensively and very successfully over the last 15 years to profile mammalian intracellular and extracellular metabolomes (e.g., HEK and CHO cells) and more recently I looked at the relationship between culture volume and agitation speed on the effect of recombinant protein production (Torres & Elvin et al, 2020, https://doi.org/10.1002/btpr.3099). Currently, the metabolite coverage is mostly focused on the major metabolites of the glycolytic pathway, TCA cycle, amino acid metabolism and nucleosides – but despite this limitation, major insights into cellular metabolism are being uncovered. However, to expand the metabolite coverage of the cell, various computational tools and open access spectral libraries are constantly being developed to improve the metabolite identification process. Much less is known about the metabolome of the various recombinant baculovirus expressing insect cell lines but hopefully with more and more metabolomics studies being performed as a matter of routine, we will see an improvement in the bioprocessing capability of this expression system in the very near future.
Since the early part of this century, many metabolic datasets have been studied to improve the bioprocessing capacity of mammalian cells (for example, in improving recombinant antibody production) and such large datasets have generated a metabolic understanding of the basic molecular principles that underpin bioprocessing – these include the metabolic pathway flux in mammalian expression systems and the relationship between cellular metabolism and the different phases of cell growth. This data has been subsequently used to improve both the design and components of chemically defined media, to optimise cell line specific feeding regimes in order to boost cellular productivity and improve recombinant protein product quality, to define metabolic markers of high cell productivity and, more recently, to define targets for specific cell line engineering. All of these things have allowed mammalian cells to produce more and more recombinant protein, moving into the grams per litre range.
Many chemically defined media and complementary feeds exist for cell culture bioprocesses and historically many of these were derived via iterative step changes from a fundamental basic understanding of the needs of a cell. Today however, with the level and depth of big datasets from the analytical power of MS, coupled with metabolic flux analysis and metabolic modelling then this allows for a much deeper understanding between specific metabolic events and bioprocess effectiveness such as protein yield and protein quality for example.
The unique precision of metabolite analysis offered by current analytical technologies allows us to address many specific questions that relate to enhanced protein manufacture. These include how to adapt cells to process scale-up and how to apply these cell processes to high-density continuous processes. The development of “on-line” process analytical technologies (PAT) that allows for the real-time monitoring of cells, their environment and the amount and quality of expressed recombinant product has been key to this analysis.
Finally, future potential lies in the development of cell line specific media and feeds that can enhance the production of cell specific recombinant proteins – for example being able to enhance the production of “novel format” or “difficult to express” proteins. If these cell specific media and feed formulations can be developed and tested to assess their impact (either positive or negative) then we could use this metabolomics based approach as a powerful strategy to improve cell productivity. This type of approach means the data could be used to create custom in-house feeds which is a potential strategy that would prove useful for smaller companies and CROs who don’t have the readily available resources to spend on extensive screening programs.
Dr Mark Elvin