How Was the Structure of G6b-B Determined?
Our journey started, as it often does, with an X-ray crystallography project which looked as if it might be relatively straightforward. The target in question was the extracellular domain (ECD) of G6b-B, a unique platelet receptor highly expressed in mature megakaryocytes and platelets. It is a type-I transmembrane protein that consists of a single N-glycosylated immunoglobulin-variable (IgV)-like domain in its extracellular region, a single transmembrane domain and a cytoplasmic tail containing an immunoreceptor tyrosine-based inhibition motif (ITIM) and an immunoreceptor tyrosine-based switch motif (ITSM).
The protein was being studied by Professor Yotis Senis and his group at the University of Birmingham who were also the first to discover it. Their challenge to Peak Proteins was firstly to determine the X-ray structure of the relatively small G6b-B ectodomain (encompassing the first 145 of the 241 amino acid full length protein) bound to a recombinant Fab fragment that they had discovered in order to identify its binding epitope. Secondly, they asked us to obtain a crystal structure of the ECD in complex with a 12 saccharide-unit heparin fragment (DP12) that was predicted to be an analogue of the physiological ligand.
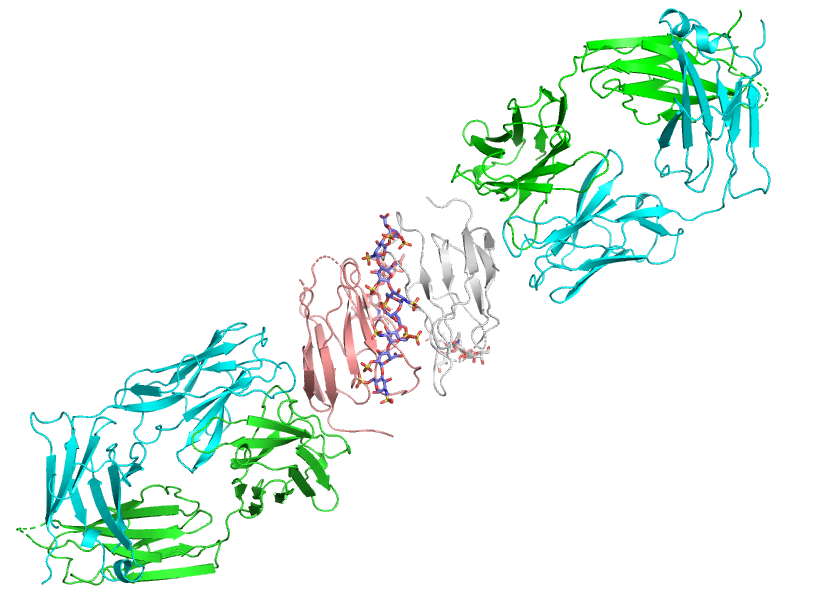
Figure 1: Ribbons representation of the Fab:G6b-B:DP12 X-ray crystal structure showing the central head-to-tail dimer of G6b-B ECD (pink & white) with a single DP12 bound at the dimer interface. The Fab fragments (green & cyan) bind to G6b-B at an epitope remote from the ligand binding site.
We started off by expressing the Fab fragment and separately a range of G6b-B ECD constructs in mammalian HEK293 cells but eventually settled on a ECD construct that spanned resides 18 to 133 which expressed well. From SDS gels and mass spectrometry analysis, however, it was clear that the purified protein was partially glycosylated at Asp32 suggesting why our initial attempts at obtaining diffracting crystals of the native construct were unsuccessful.
Mutating the offending asparagine residue to an aspartate successfully removed the N-glycosylation but further attempts to crystallise the mutated ECD either both alone or in complex with DP12 were still unsuccessful. We were therefore pleasantly surprised when 3 months later we discovered that crystals of the G6b-Fab-DP12 complex had grown slowly over this period – although frustratingly they only diffracted X-rays to less than 4 Ångstroms resolution.
Although this data was sufficient for us to locate the positions of the two Fab molecules in the crystal by molecular replacement it was not enough to clearly resolve the unknown structure of the G6b-B ECD itself. Tantalizingly, however, limited regions of unmodelled electron density were observed close to the complementarity-determining regions (CDR) of the Fabs and were thought to be of the bound G6b-B.
Further investigation of the mutant G6b-B construct with intact mass spectrometry unexpectedly revealed that its measured mass was 948 Da greater than predicted, a mass consistent with the protein also possessing a commonly found O-linked oligosaccharide. This O-glycosylation was subsequently located using TOF-mass spectrometry of a chymotryptic digest to a region encompassing residues 66–80 which contained five potential modification sites. Not knowing which of these residues were responsible for the glycosylation each were individually mutated to Ala in tandem with the N32D mutation.
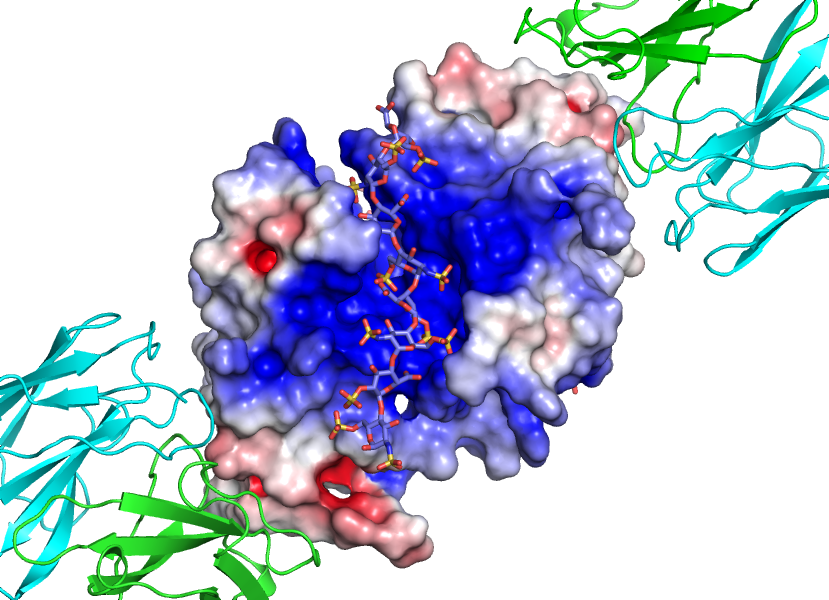
Figure 2: Closer view of G6b-B dimer with superimposed electrostatic potential surface showing heparin ligand (DP12) positioned in a interdomain cleft with exquisite shape and charge complimentarity.
Subsequent analysis of all of these mutants clearly showed that the predominant O-glycosylation site was on Thr73, although the other four residues could also be modified to a lesser extent. Using this information, a further two constructs were generated and tested for crystallization, one with the four other residues mutated to Ala but leaving Thr73 intact and the second with all five residues mutated.
The former construct was soon found to crystallize readily as a G6b-Fab-DP12 complex and allowed X-ray diffraction data to be collected to 3.1 Ång at the Diamond Light Source synchrotron near Oxford. With this improved resolution we were now able to iteratively build and refine a model encompassing about 90% (101 out of 116 residues) of the of two G6b-B ECD chains found in the crystal. Excitingly at this stage the residual electron density revealed the presence of a single molecule of the DP12 ligand bound to the ECDs with good density covering 8 of the 12 saccharide units.
The final refined model represents a complex of G6b-B ECD, DP12 and Fab fragment chains in the ratio 2:1:2, respectively. This structure reveals that the two G6b-B ECDs form a head-to-tail dimer and that the DP12 heparin ligand binds with exquisite shape and charge complimentary to a cleft created at the dimer interface remote from the Fab binding epitope.
Size-exclusion chromatography was also able to show that ECD dimer formation only occurs in solution as a result of heparin binding. This one structure therefore, although not straightforward to obtain and requiring great skill and determination by our crystallisation team, was able to successfully achieve all the objectives set by the client, to identify the epitope for the Fab fragment and also for the first time reveal the details of ligand binding to this important platelet receptor.
As Professor Yotis said “Peak Proteins doggedly persevered from start-to-finish, achieving our objective more efficiently and economically than would have been possible otherwise”. It certainly required crystallography perseverance and protein mass spectrometry detective work to crack the case, but the results certainly made the journey very worthwhile!
Further details are described in the publication : Heparan sulfates are critical regulators of the inhibitory megakaryocyte-platelet receptor G6b-B. Vögtle T, et al. Elife. 8. pii: e46840. 2019.


