Adapting use of size exclusion chromatography to improve yields of an unstable, purified protein
A client asked us to express and purify at least 2 mg each of two similar enzymes for selectivity assays. There was literature precedent for both proteins, and a similar approach would be taken with each, purifying via affinity chromatography followed by size exclusion chromatography. However the literature referred to one of the proteins as having some stability issues.
One protein expressed well and we were able to generate >2 mg of pure monomeric protein with good activity. The other (as described in the literature) was unstable when running the purification at scale, giving unreasonable losses when concentrating the protein, and did not elute as expected from the size exclusion column. Time was also key as the protein tended to aggregate as the purification progressed resulting in loss of activity.
Our Solution
We had to completely re-think the purification steps from start to finish in order to minimise time delays and eliminate the potential for losses. The affinity capture step worked reasonably well, but had to be optimised to maximise binding to the resin, and minimise elution volumes. This avoided the need for concentration (and the associated losses) prior to the size exclusion step. However, on larger size exclusion columns the protein was unnaturally retarded, and the yield was significantly less than expected. Moving to a smaller column length, increasing the salt concentration to counter-act ionic interactions, and doing shorter/faster runs allowed more of the protein to elute in an active, monomeric state. This set up required smaller load volumes, but this was accommodated by having the smaller elution volumes off the affinity resin.
The proportion of aggregated to monomeric protein increased with time (seen over a 6 hour period), as was clearly observed on the elution profiles from the size exclusion column (see UV elution profile below).
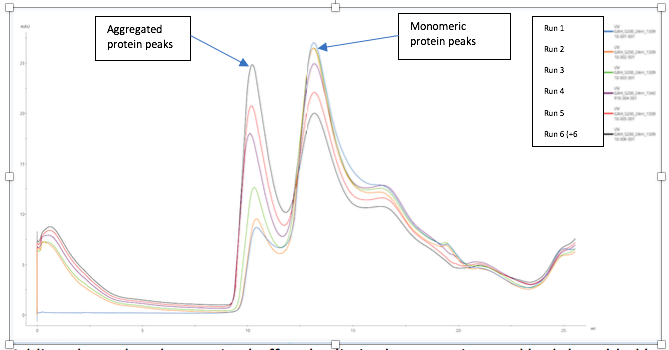
Adding glycerol to the running buffer also limited aggregation, and had the added bonus of stabilising the protein sample during the freeze/thaw process, allowing the protein to retain its monomeric state and good biological activity. This was confirmed by analytical size exclusion chromatography and activity assays.
The Impact
These modifications to the purification process made it possible to address the reported stability issues, taking the protein from the cell lysis stage through to freezing over the course of a working day. Having streamlined the process, and minimised losses due to aggregation, we were able to provide the required 2 mg of this protein to the client, in a monomeric, and active state.


