Identification of Novel Post Translational Modifications of a Recombinantly Expressed Peptide
A client had requested to obtain a small peptide that was <10kDa, normally processed by cleavage of a larger precursor protein, to be supplied as an untagged recombinant protein. At Peak Proteins, we have experience working with expressing and purifying this type of peptide. Here we show how both construct design and careful planning for an effective purification method and critically, extensive characterisation of the peptide, resulted in an enhanced data package and protein production that exceeded the client’s request.
Our Solution
Smaller proteins or large peptides can be difficult to express, however this problem can usually be solved by the addition of fusion proteins. The construct we created included the fusion protein, maltose binding protein (MBP) along with 6His and a TEV cleavage site. The use of the MBP allowed the peptide to be successfully secreted in large amounts from our HEK293 suspension cells expression system, where cells are grown in serum-free media (Figure 1). After capturing the protein via nickel affinity chromatography, the MBP was cleaved from the peptide by using TEV protease which cleaves at the TEV cleavage sequence (ENLYFQ/G). In this case the client could not accept an additional Gly at the N-terminus so we butted the ENLYFQ sequence against the desired N-terminal residue which should have been amenable to TEV cleavage. Removal of any uncleaved protein as well as the TEV protease (which is also 6His tagged) was achieved through a second subtractive nickel column (Figure 2). This protocol worked incredibly well and resulted in >90% pure untagged protein, which the client had requested.
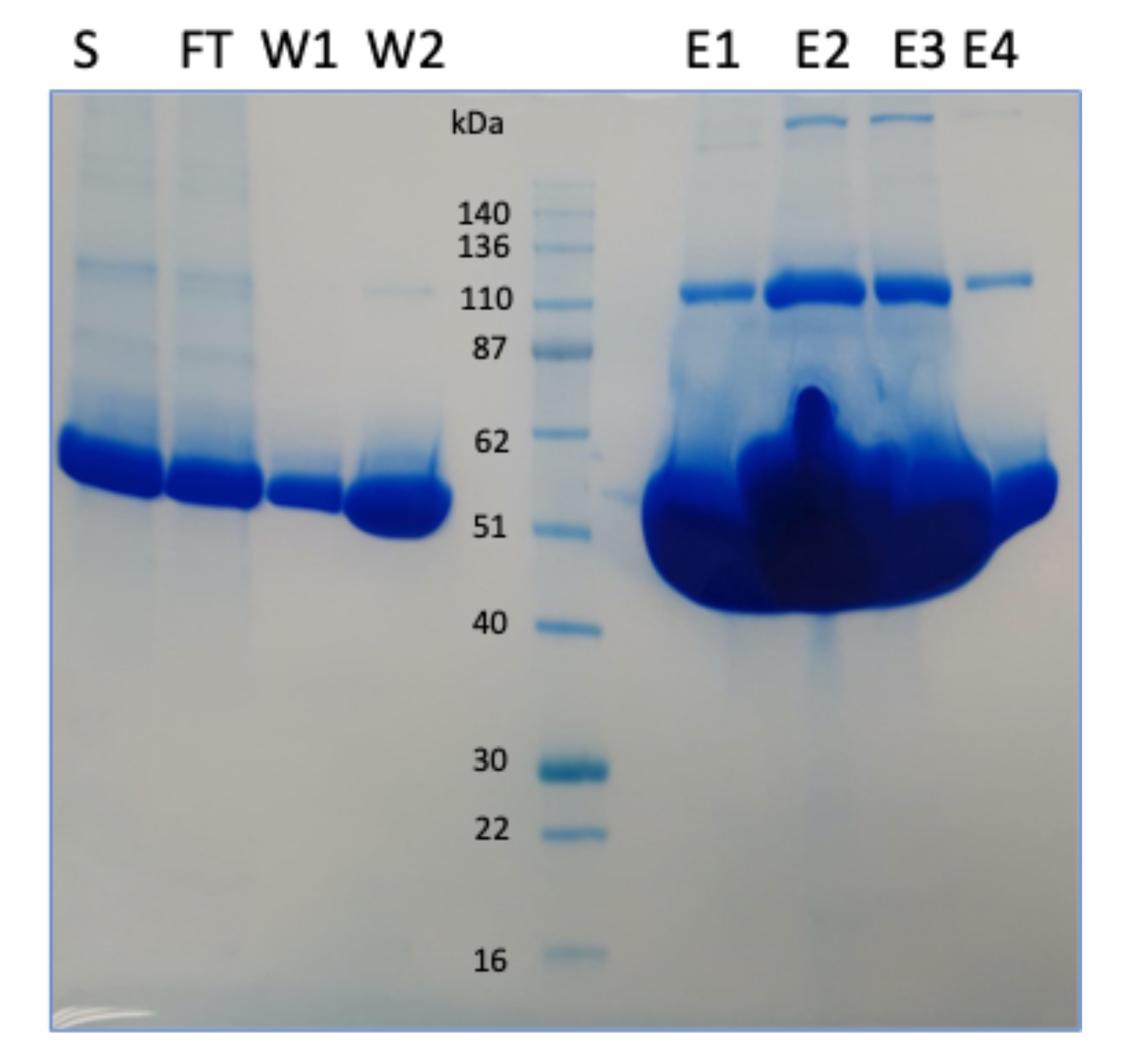
Figure 1: SDS-PAGE gel showing the elutions after nickel affinity chromatography; S-supernatant, FT- flowthrough, W-washes, E-elution

Figure 2: SDS-PAGE gel showing the protein pre and post TEV cleavage
As part of our protein quality check, we routinely analyse proteins using our in-house mass spectrometer. The intact mass spectrum revealed some very unexpected yet interesting results regarding post translational modifications. This specific protein was revealed to be O-glycosylated; identified with a mass addition of +947Da (Figure 3). This corresponds to the mass of an O-glycosylation – Hex(1)HexNAc(1)NeuAc(2). In addition to the 947Da mass, other peaks are seen that indicate the presence of HexNAc, hexose and sialic acids that may be observed due to in-source fragmentation. These would all be consistent with a commonly observed O-linked tetrasaccharide (Vögtle et al., 2019). As well as this glycosylation, the intact mass trace also revealed that this protein had 3 disulphide bonds (as seen by the deduction of -6Da). These modifications were previously unreported in both the literature and Uniprot, and clearly demonstrated the benefit of conducting thorough intact mass spectrometry analysis.
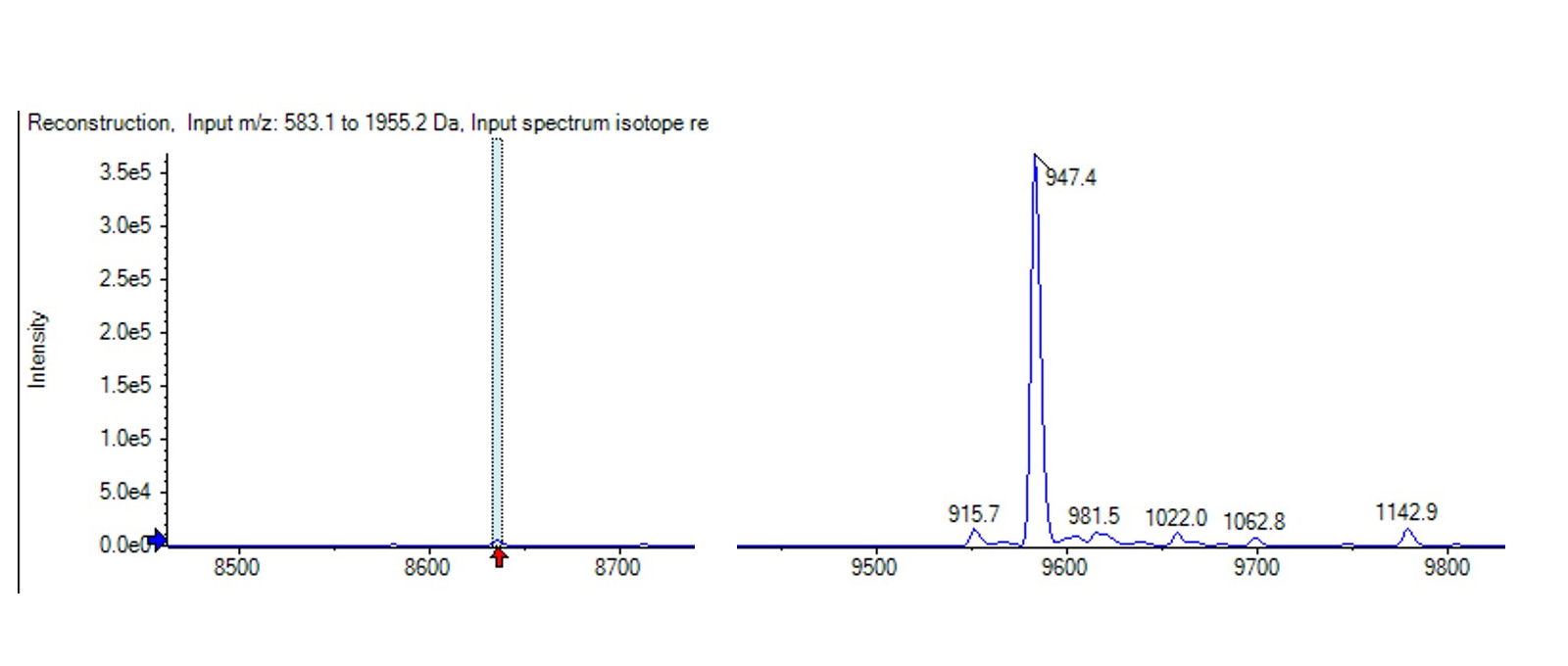
Figure 3: Intact mass spectrometry spectrum showing the peak at +947 which indicates O- glycosylation
The Impact
The careful planning of this purification, from construct design through to purification showed how useful the MBP fusion can be in driving the expression and secretion of small peptides from mammalian cells. Moreover, this case study demonstrates how our protein characterisation services using mass spectrometry are extremely useful in identifying any post translational modifications your protein may possess.


