Mapping N and O glycosylation sites using mass spectrometry
Post translational modifications (PTM) play a key part in protein function. One of these is glycosylation, the addition of sugar molecules to a protein by a covalent bond. There are two kinds of glycosylation seen in proteins: N-linked glycosylation and O-linked glycosylation. A further explanation of these can be found in our previous blog post.
Sometimes, when purifying a protein for the first time, unexpected findings can crop up. In this case, where one protein band was expected, we saw double banding.
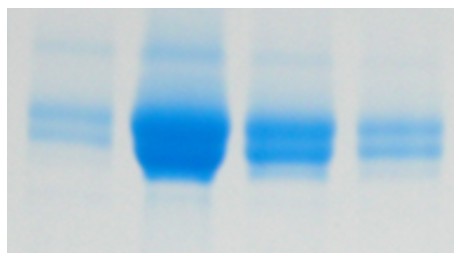
Double banding can occur for a number of reasons. Sometimes, it’s as simple as a prominent contaminant protein of a similar weight as the target protein. If that is not the case, we then look for other explanations such as has proteolytic degradation occurred or is there PTM of the target protein?
For this project the two bands were sent for peptide mapping. It was found that both bands contained the target protein, therefore we needed to dig deeper for an explanation.
When peptide mapping, a ‘top-down’ approach is often used when looking at PTM; the specific modification has to be inserted into the software so that it can search for peptides containing the exact modification. When the modification is not known, there are a selection of commonly seen modifications that can be searched for. Intact mass data can also be used to decide which modifications to search for, as we can see how they affect the overall mass of the protein.
In this case, as the protein is known to be glycosylated, it was assumed that this was cause of the double banding. To look into this we incubated the protein with Endo H. Endo H is a specific enzyme which hydrolyses the bond between two GlcNAc molecules in an N-linked sugar. This leaves behind N-acetylglucosamine. After the hydrolysis reaction was taken to completion, it was expected that the double banding would no longer be present. However, when an SDS-PAGE gel was run, the double banding was still seen, implying that there was a different cause.
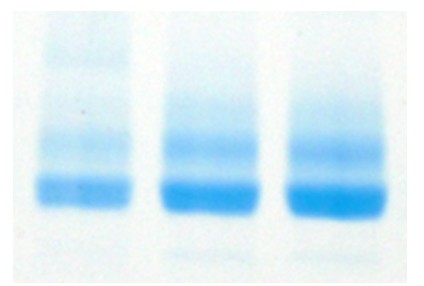
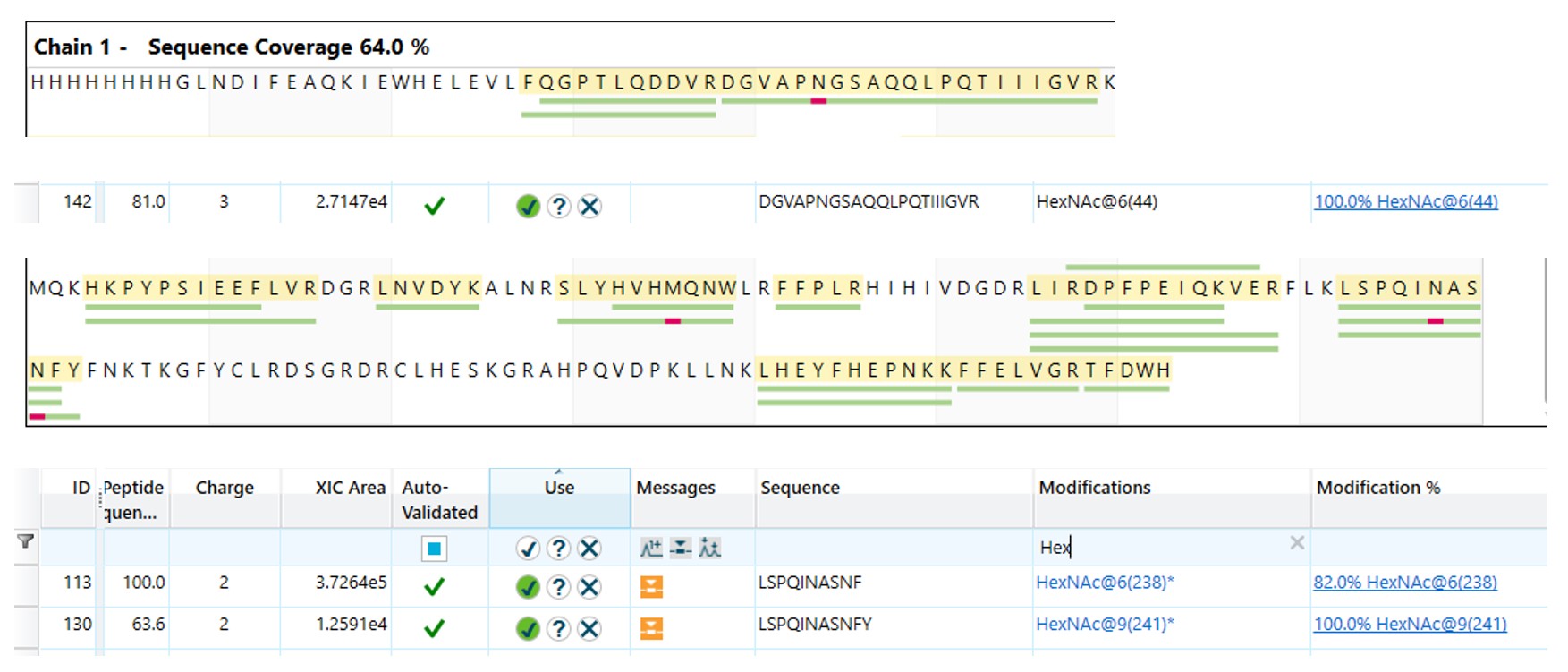
When the Endo H treated protein was run on intact mass, some familiar mass additions were seen.
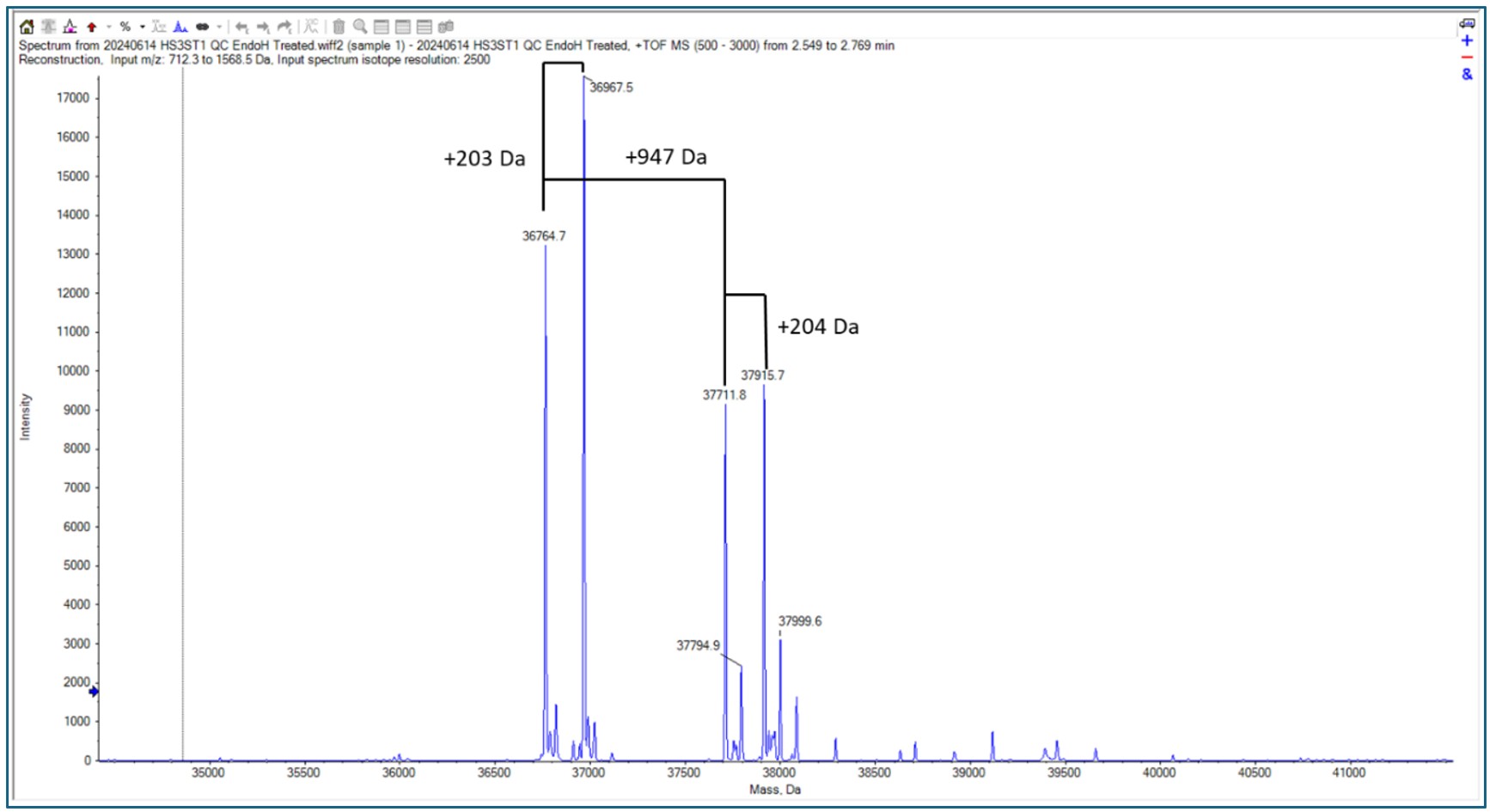
A 203 Da and 947 Da addition are indicative of glycosylation PTMs, specifically N-acetylhexosamine and Hex(1)HexNAc(1)NeuAc(2) respectively. Therefore, these modifications were searched for against the peptide mapping data.
One modification was only seen in the top band and not the bottom band. This was a Hex(1)HexNAc(1)NeuAc(2) on Thr(36). As this O-glycosylation was only seen in the top band, it is likely that this was the cause of the double banding.
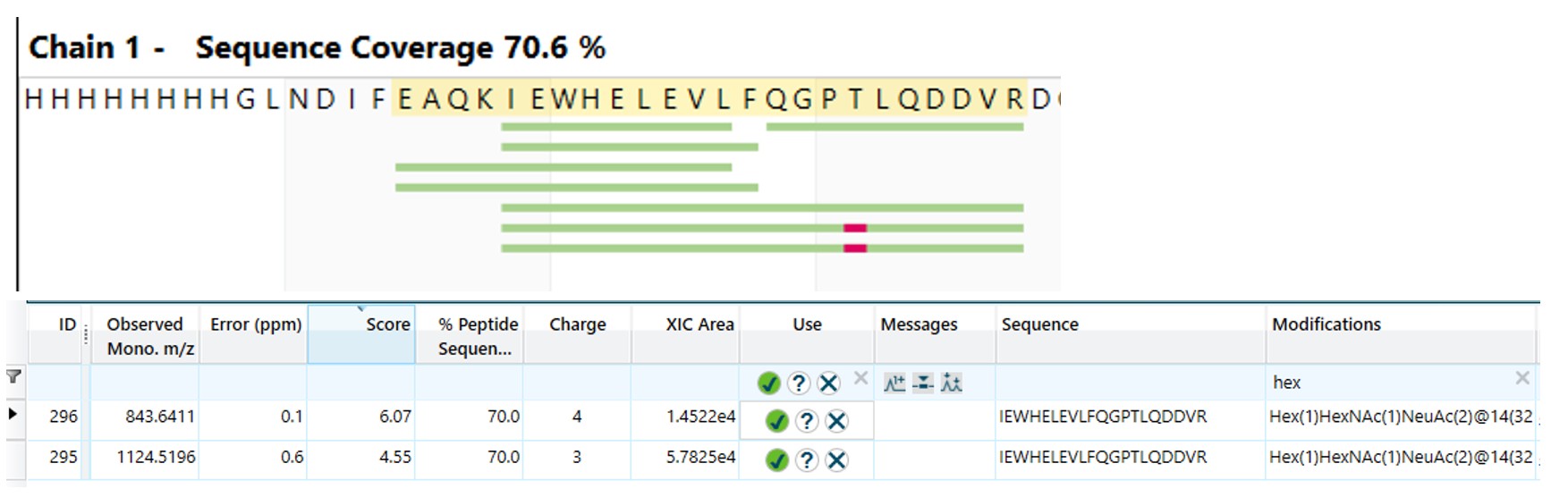
The N-glycosylation sites on the protein were going to be shown by the N-acetylhexosamine left behind by the EndoH hydrolysis. These were found on Asn(48), Asn(242) and Asn(245).
Conclusion
By using peptide mapping and intact mass analysis, we were able to map three N-glycosylation sites and one O-glycosylation site on the target protein.


