More Papain, More Problems
The Challenge
A client asked us to generate Fab and Fc pools from a full-length antibody using Papain digestion. In this case study, Camila Torres, one of our protein scientists describes the issues that we encountered and the conclusion we came to for tackling this type of request in the future.
Papain is a cysteine protease which can be used to generate Fab and Fc fragments from full-length antibodies. It is, however, a very promiscuous protease and can cleave at many different residues. Our concern was that this can sometimes lead to issues after its use and can make reproducing a protocol difficult for subsequent batches.
Initial Results
After papain digestion of the antibody, the Fab and Fc fractions were separated using a protein A column.
However, a size exclusion run of the isolated Fab fraction showed a very peculiar result: instead of a single Fab peak, there were two elution peaks.
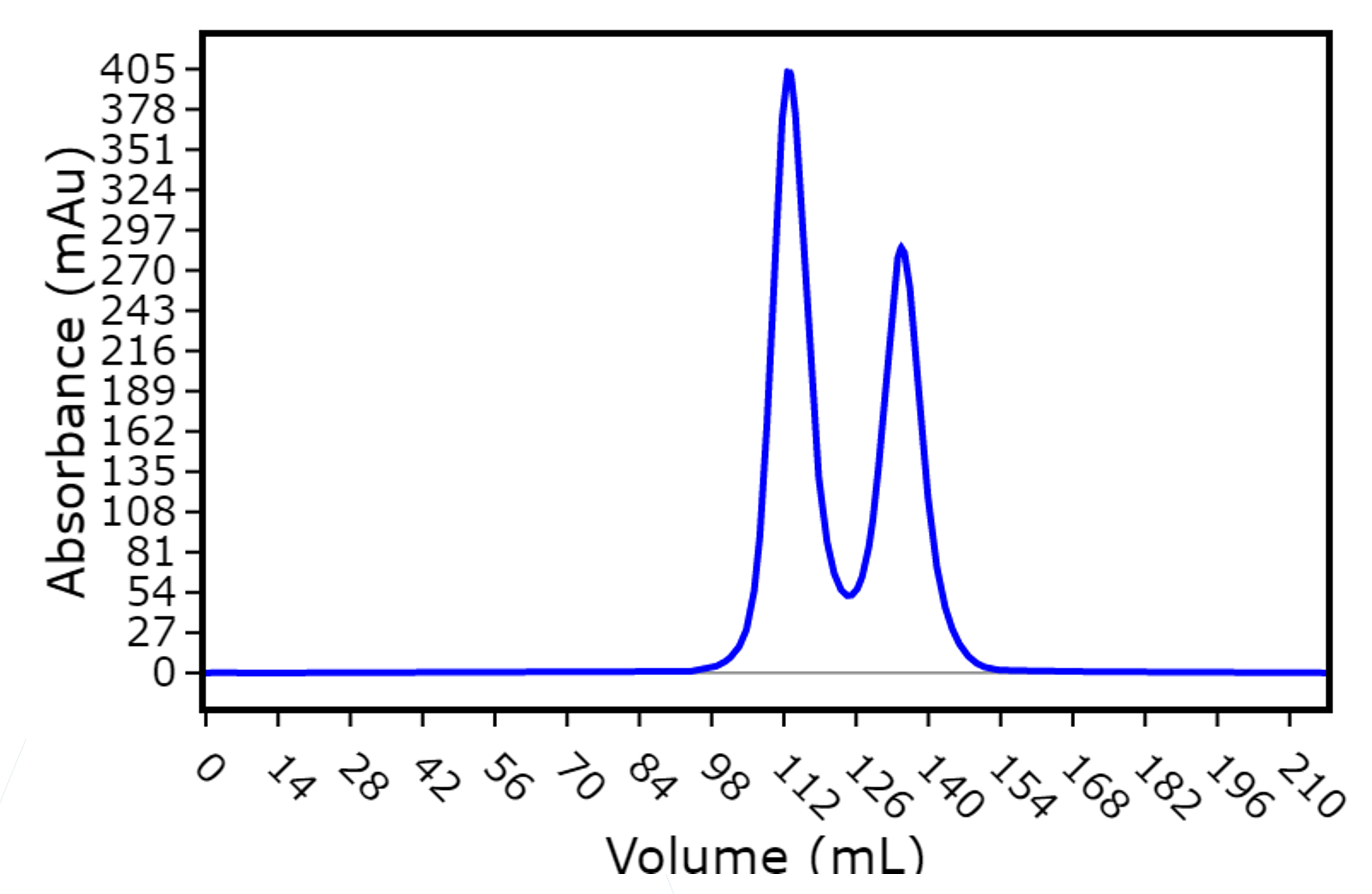
Figure 1: Fab SEC elution from a 180 mL 16.60 S200 column
A non-reduced SDS-PAGE gel of the elution peaks gave no further insight.
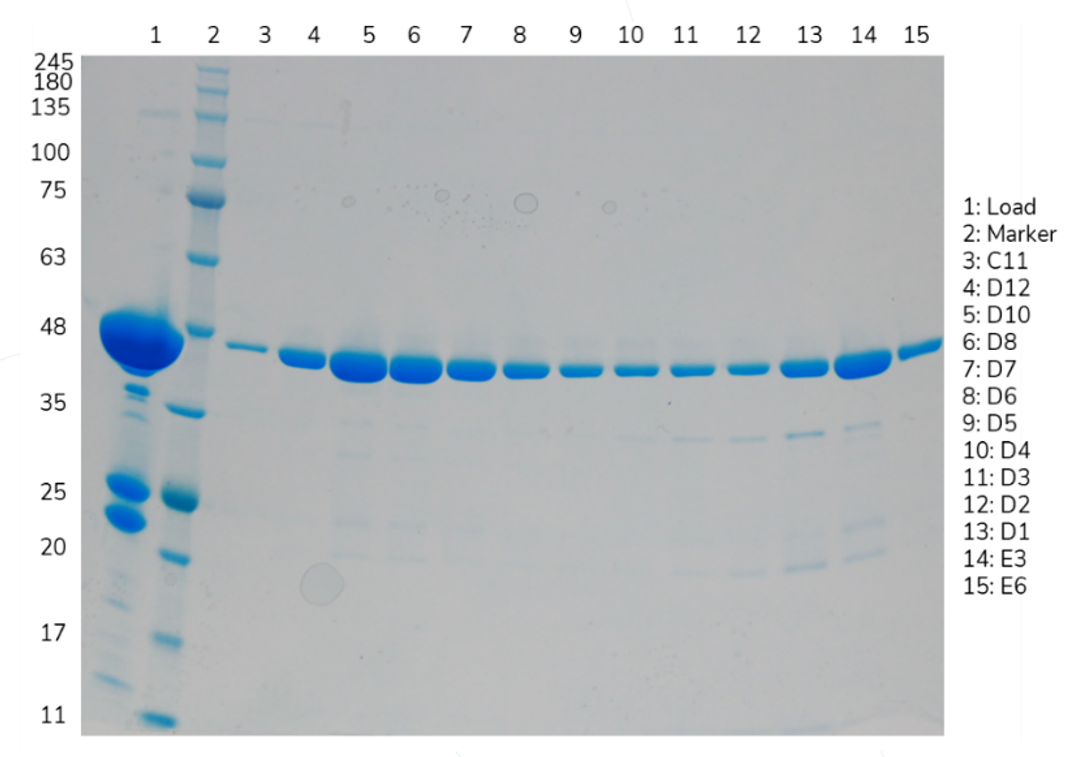
Figure 2: Non-reduced SDS-PAGE gel of the Fab SEC elution
Both peaks were shown to contain the Fab, with minor contaminants seen in the second peak.
However, a reduced SDS-PAGE gel of the elution peaks just raised further questions.
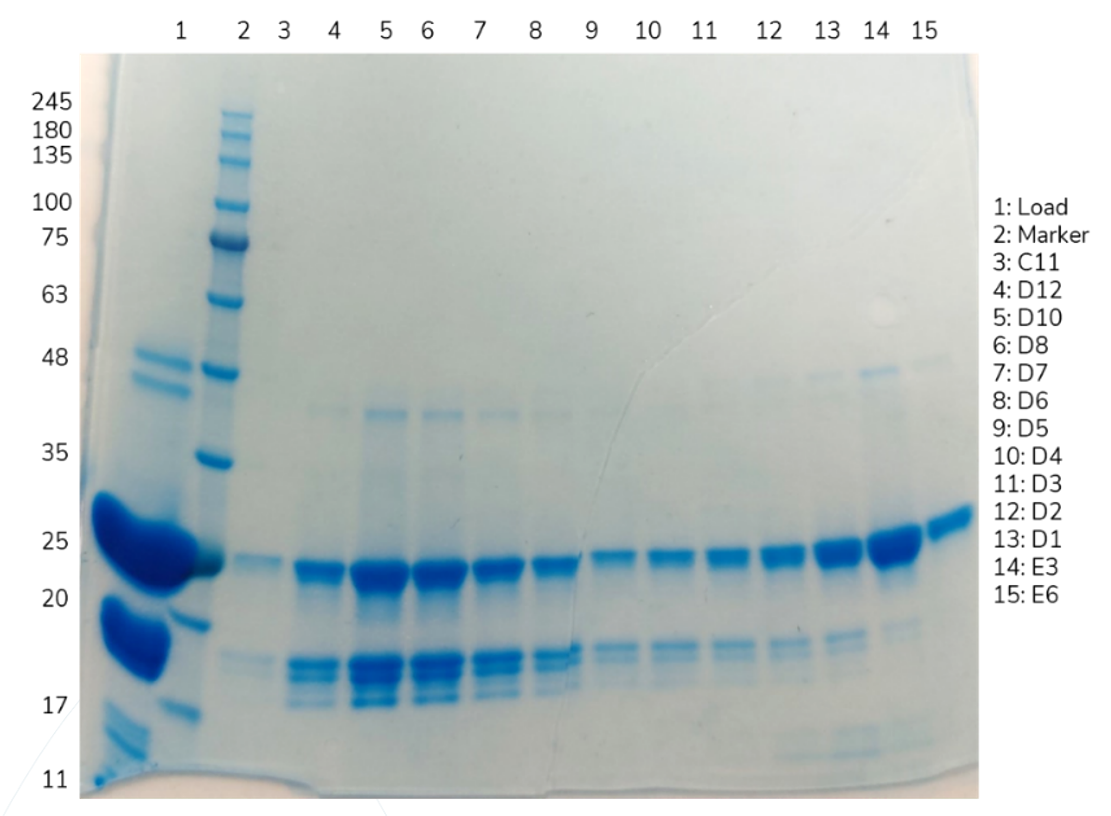
Figure 3: Reduced SDS-PAGE gel of the Fab SEC elution
The bands at 25 kDa corresponded to the heavy and light chains of the Fab, separated due to the reduction of the disulfide bonds which held them together. But what were the bands between 17 and 20 kDa?
The two peaks were pooled and concentrated separately. They were then run on intact mass to try to gather further information.
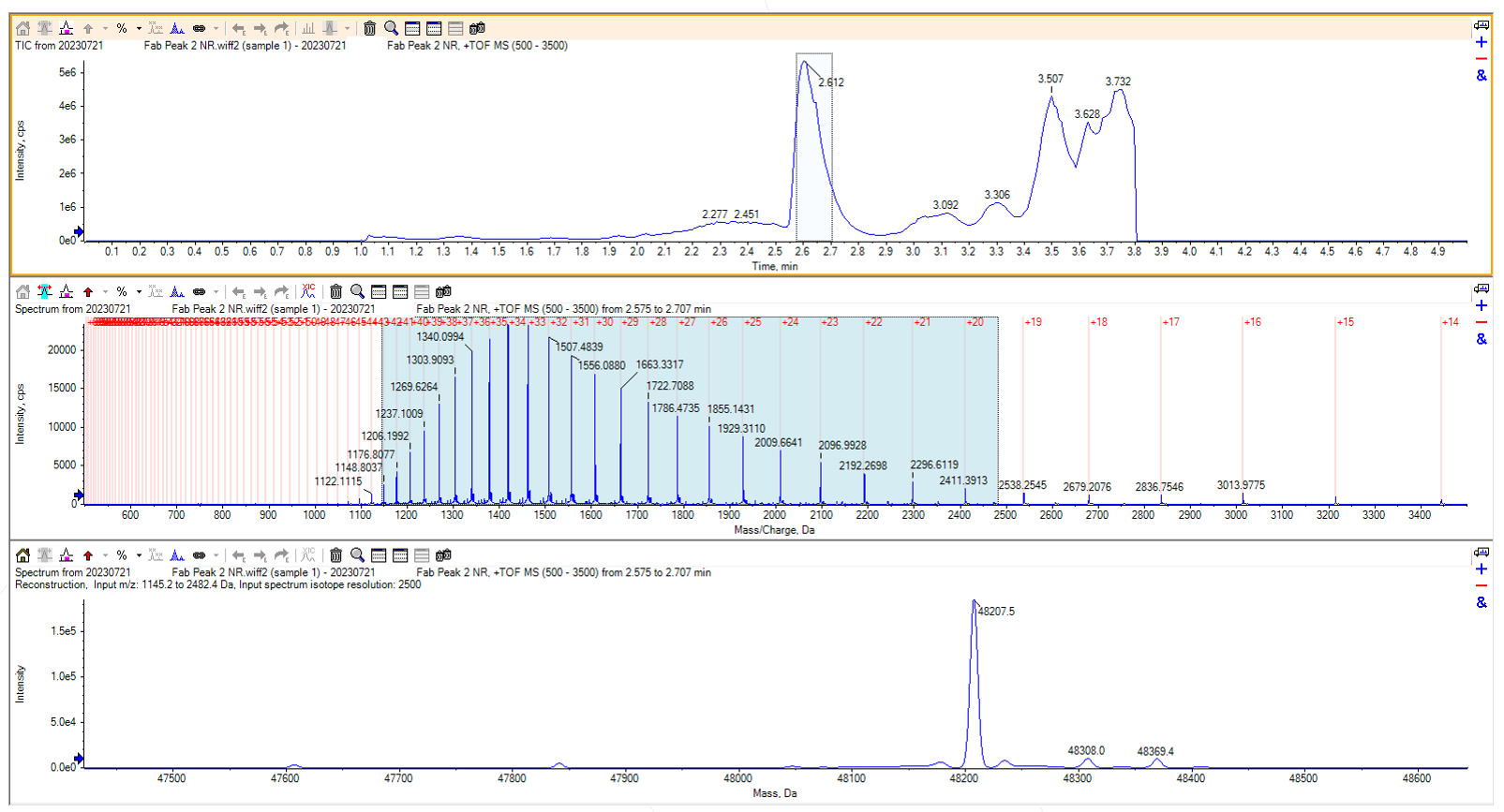
Figure 4: Non-reduced intact mass chromatogram of the second Fab peak
Analysing the chromatogram from the intact mass run of the second peak was quite straightforward; the main peak at 48207.5 Da matched the expected mass of the Fab, minus 10 Da from the five disulfide bonds within the Fab.
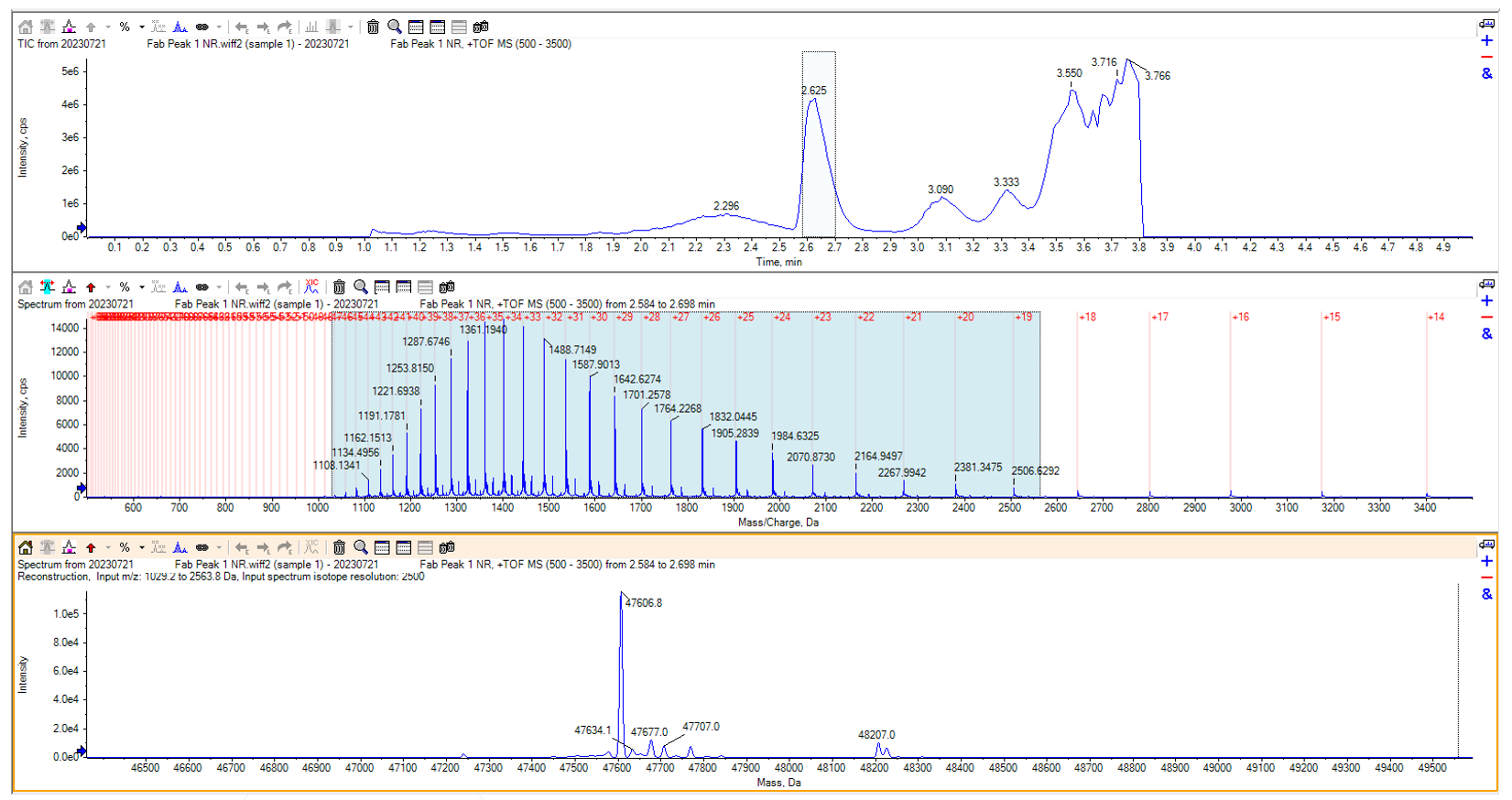
Figure 5: Non-reduced intact mass chromatogram of the first Fab peak
Analysing the chromatogram from the intact mass run of the first peak was more difficult. The main peak shows a mass of 47606.8 Da. This – 600.7 Da difference did not match any expected modifications, nor did it match an N-terminal or C-terminal truncation on either chain. Further investigation needed to be done.
A small sample from the first peak was reduced with DTT and heated at 70 °C for half an hour to see which insights could be gained from separating the light and heavy chains.

Figure 6: Reduced intact mass chromatogram of the first Fab peak
The main peak of 23450.7 Da corresponded to the light chain of the Fab and the much smaller peak of 24766.1 Da corresponded to the heavy chain of the Fab.
The other peaks, 18258.1 Da and 5907.1 Da, were more difficult to assign. Once again, the masses shown did not match either an N-terminal or C-terminal truncation of the heavy chain. Furthermore, adding up the masses of two peaks gives a mass of 24165.2 Da which is too small to be the heavy chain.
After some trial and error, it was discovered that two papain cleavages in the middle of the heavy chain resulted in two fragments of 18.3 kDa and 5.9 kDa being formed, plus a six-amino acid fragment, GEPTYA, which could not be seen in the intact mass chromatogram.
Adding up the 18258 Da fragment, the 5907 Da fragment and the 23450 Da light chain, gives a mass of 47615 Da. This, minus 10 Da from 5 disulfide bonds, matched the main peak in Figure 5, of the non-reduced sample from peak 1.
The 18.3 kDa fragment and 5.9 kDa fragment could not be seen in the non-reduced SDS-PAGE gel (Figure 2) because the intramolecular disulfide bonds between different cystine residues in the heavy chain kept the three fragments together; once the disulfide bonds were reduced, the fragments were no longer kept together and so the fragments could be seen in the reduced SDS-PAGE gel (Figure 3).
Conclusion
This papain over-digestion is a common issue which is difficult to correct. The line between under-digestion and over-digestion when using papain is not so much a fine line as an overlapping, fuzzy area – here, the digestion of the immunoglobulin was not taken to completion, but over-digestion of the Fab fragments still occurred.
Our Recommendation for Producing Fabs for Future Projects
One way to circumvent this issue, is to produce the Fab as a recombinant protein using mammalian expression systems and we have several examples where we have done this for our clients. This removes the issue of papain altogether and usually leads to a higher Fab yield. Of course this approach does rely on having the target amino acid sequence of the Fab in question, but assuming that is available, our recommendation is for a recombinant approach to produce Fabs.


