Sherlock Holmes and the Case of the Three Missing Amino Acids
A client recently requested the production of two bispecific antibodies and provided us with the heavy and light chain variable sequences that they required. After reviewing the literature, we decided to produce these bispecific antibodies as fused, single-chain variable fragments (scFvs) with an N-terminal leader sequence to enable secretion from mammalian cells and a C-terminal affinity tag to facilitate purification.
The two scFvs were fused genetically to generate a single polypeptide chain in the following format:
Signal peptide-(Ab1 Fv light-linker-Fv heavy)-linker-(Ab2 Fv heavy-linker-Fv light)-affinity tag
We were also asked to make each of the antibodies as monovalent scFvs to use as controls and opted to keep the same combination of heavy and light chains as represented in their equivalent bispecific format. Both bispecific molecules expressed and purified well whereas one of the scFvs did not express at all (see below).
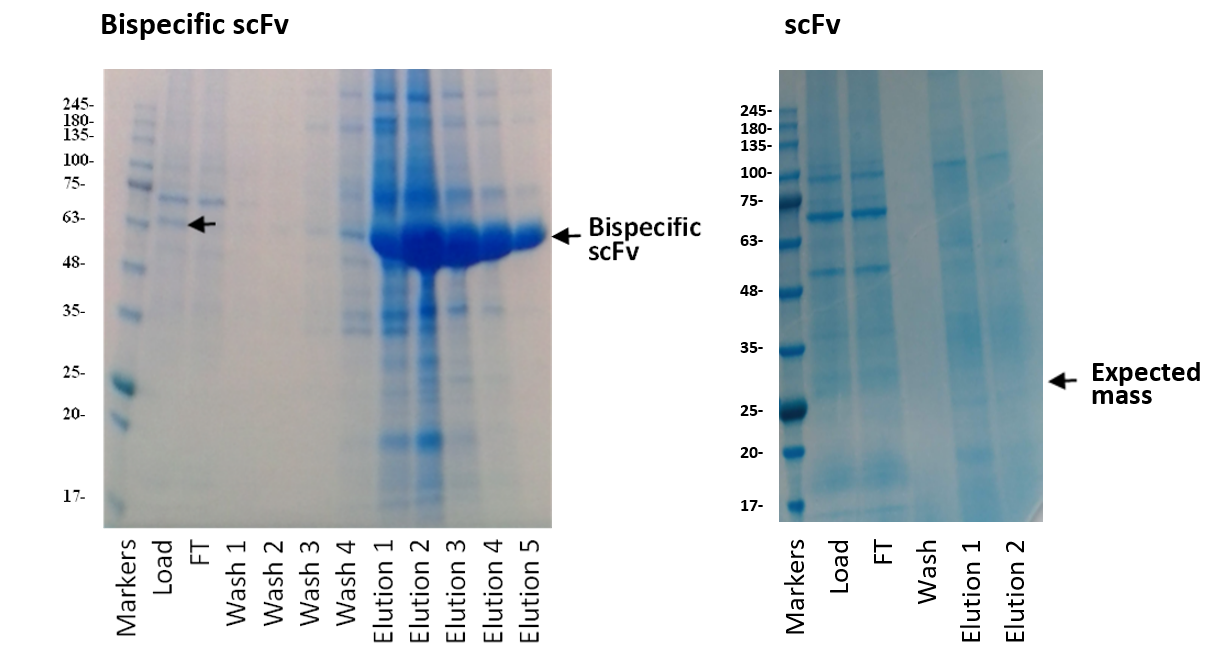
We were surprised as to why this had happened when the same sequence had expressed extremely well as a bispecific scFv and could even be seen in the supernatant pre-purification. By aligning the provided sequence against the database, we noticed that this sequence omitted the three amino acids EVQ at the N-terminus and instead started with a leucine. Furthermore, visualisation of antibody 3D structures showed that the sequence EVQ at the N-terminus of heavy chain is visible and becomes part of a beta strand.
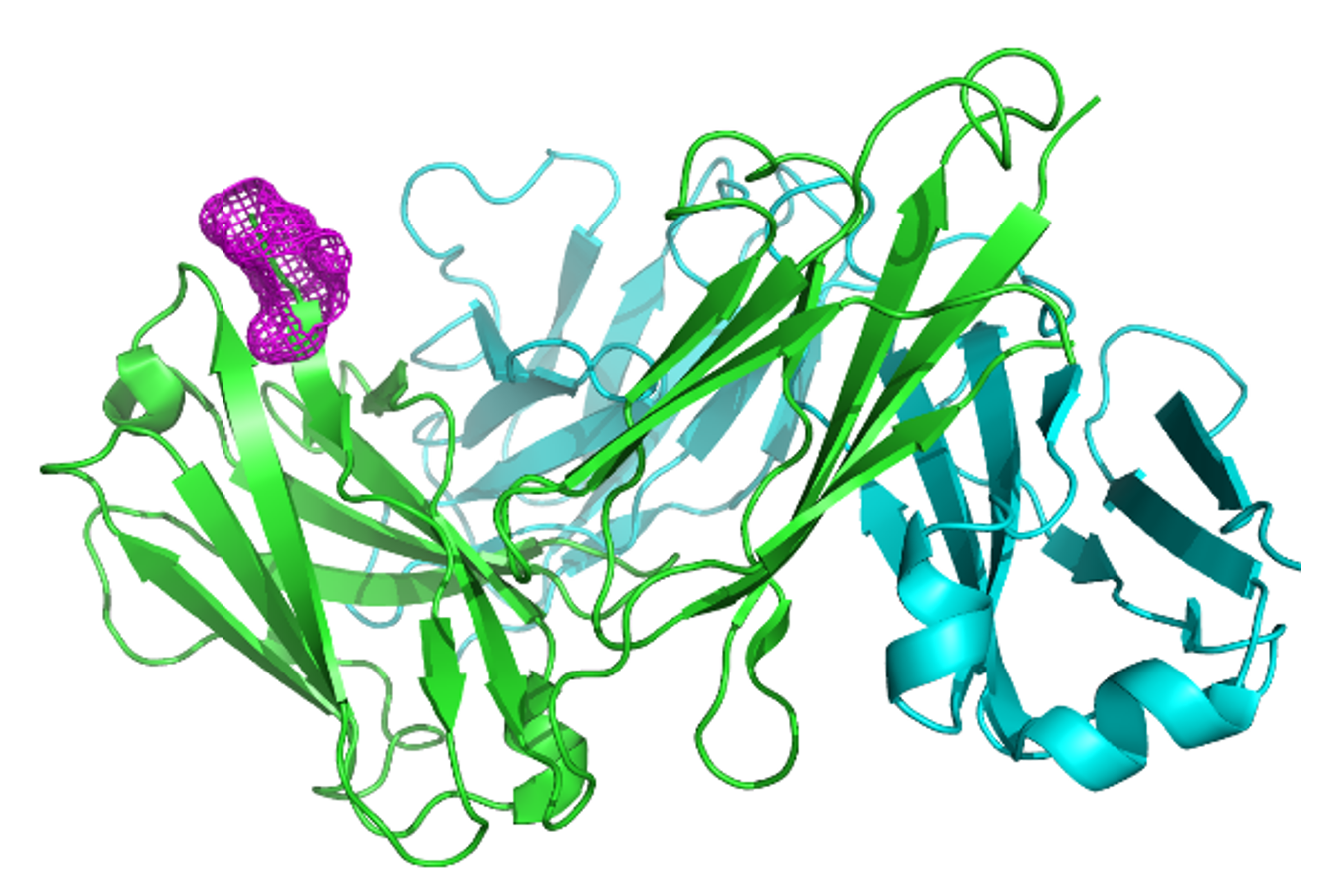
Figure 2: A Fab with EVQ highlighted in pink
We hypothesised that when these three amino acids were missing it destabilised the whole antibody. The EVQ amino acids that had been omitted were added in front of the leucine and the scFv construct re-expressed. As shown in the figure below, the modified scFv expressed very well and was straightforward to purify. We think the reason the truncated sequence may have expressed well in the bispecific molecule is because it was preceded by a GGGGS linker rather than following on immediately after the signal sequence. Case solved!
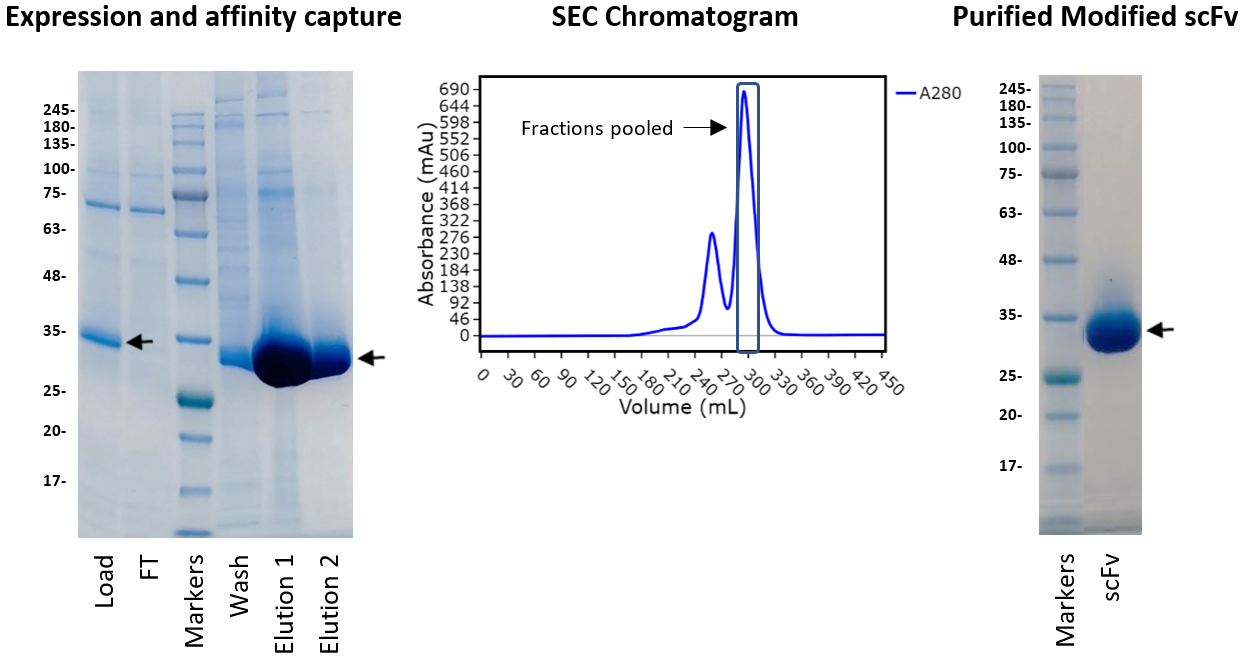
Figure 3: PAGE showing purification samples of the amended scFv


