Use of mass spectrometry to elucidate critical quality attributes of a biopharmaceutical
In this case study Tomas Adomavicius discusses a project where we employed various mass spectrometry techniques to confirm the target API was as expected and also to elucidate the nature of two impurity peaks that were observed during the development of a downstream process for a biopharmaceutical.
Background
The early identification of critical quality attributes (CQAs) [1] in the development of a biopharmaceutical production process is key to delivering a “Quality By Design” [2] based submission package to the regulatory agencies. In particular, in-depth knowledge of the CQAs, with assays to measure them, allows for the development of processes that are robust, reproducible and therefore amenable for full scale manufacture.
Our customer is developing upstream and downstream production (USP and DSP) processes for a biopharmaceutical. The ultimate Active Pharmaceutical Ingredient (API) is a recombinant protein that consists of two disulphide-linked peptide chains, which then has a compound covalently linked to a specific residue on one of the chains (Fig 1).
The challenge
During their DSP experiments two contaminant peaks were observed in analytical RP-HPLC. The request was to confirm the identity of the main API peak and also to try and find out the exact nature of the 2 contaminant peaks. They collected fractions from these peaks and shipped them to us to perform mass spectrometry analysis.

Figure 1. Diagram of the target API: Two disulphide linked peptide chains (purple and teal) with a compound (red) covalently linked to a specific residue on one chain.
Confirmation of the API product
Intact mass spectrometry analysis of the sample from the RP-HPLC that was suspected to be the main target API peak gave a single mass that was exactly as predicted from the amino acid sequence plus the linked compound. Furthermore, no significant additional peaks were observed that would suggest any post-translational modifications such as oxidation.
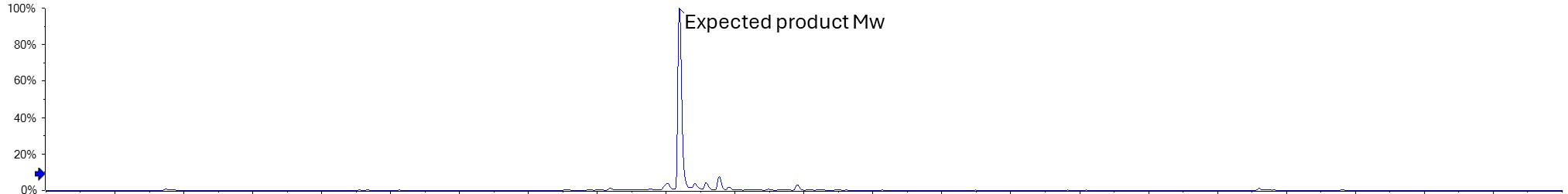
Figure 2. Deconvoluted intact mass of the biopharmaceutical, showing the exact mass as expected
Peptide mapping then confirmed the sequence was as expected and we were also able to see the peptides with the compound bound to the expected residue. The data obtained was also challenged to see if any post translational modifications were evident and none were identified. This was the first conclusive data the customer had that confirmed the correct identity of their target API molecule.
Identification of contaminant peak 1
For the first of the contamination peak samples, intact mass was the same as main product. However, reducing intact mass gave two peaks and suggested that the compound was present on the wrong peptide chain. The initial peptide mapping experiment we ran didn’t give us a peptide with the compound bound.
When we looked in more detail into the sequence and the data from the main API peak peptide mapping, it was eviden that binding of compound increases the hydrophobicity of the peptide, and the alternative binding sites are located on more hydrophobic peptides, so it was possible that the new compound-bound peptides would bind too tightly to the RP-HPLC column that is upstream on the mass spectrometer instrument. By adjusting the elution gradient we were able to see this peptide in a subsequent peptide mapping run and were able to confirm that the compound in this sample was bound to the N -terminus of the peptide chain 1. (Fig 3)
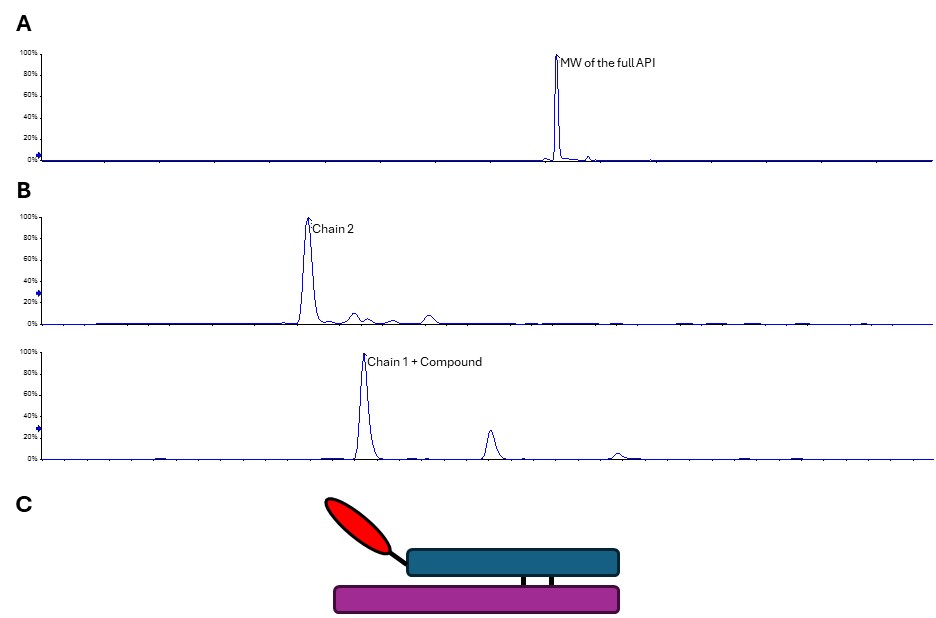
Figure 3. Configuration of contaminant peak 1. A. Deconvoluted intact mass showing the same MW as the main API sample. B. Reduced sample splits into two peaks, one matching to the expected mass of Chain 2, and the other to Chain 1 + Compound. C. Model of the contaminant, with the compound bound in the wrong place on the N-terminus of peptide chain 1
Identification of contaminant peak 2
For the second of the contamination peak samples, we found no match to the expected intact mass. However, it did match to a protein dimer, plus one compound, minus 18 Da. It appeared that a side group of the compound is reactive and can form a covalent bond with a separate protein molecule, losing H2O. We therefore hypothesised that we are observing a compound-linked protein dimer. (Fig 3). Furthermore, reducing intact mass showed that the dimer is linked via chain 2, as we observed masses for Chain 1 and Chain2-CPD-Chain2 (-18 Da) (Fig 4).
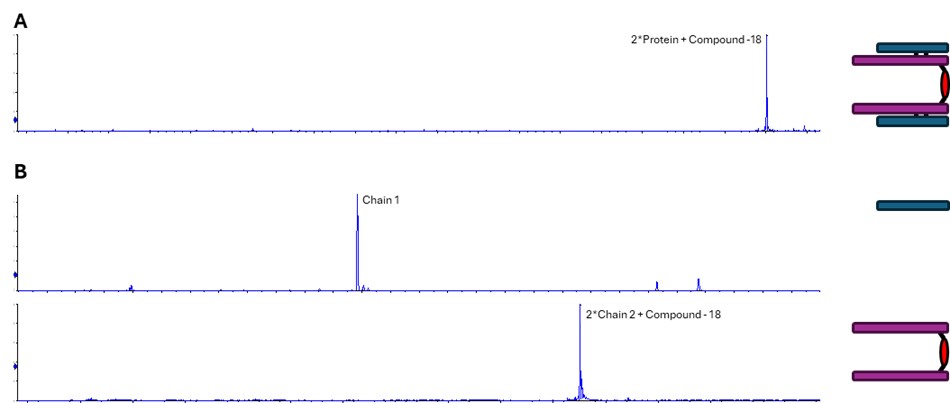
Figure 4. Confirmation of the contaminant peak 2 as observed in non-reducing (A) and reducing (B) intact mass analysis. A dimeric form of the API has been formed by the compound binding to chain 2 of two API molecules.
For completion, during all of the peptide mapping experiments we searched the data against the Swissprot database to check for the presence of host cell proteins (HCPs) that may have been carried over from the USP. None were observed, helping to rule out the potential CQA.
Impact
We used a combination of reduced and non-reduced intact mass along with peptide mapping analysis to give our customer their first data that confirmed the biopharmaceutical API molecule was as expected. Obviously, this is critical data to ultimately support their regulatory submission. In the short term is has confirmed for them which peak on their RP-HPLC analysis is the target API molecule to track during their process development experiments where they aim to improve both purity and yield of the API.
We were also able to identify the structure of the two main contaminants they observe during their DSP. This data now allows them to monitor these contaminants and help them refine the process to reduce the levels of these in the final product. We were also able to show that no post translational modification of the API molecule had occurred.
Overall, this mass spectrometry data has provided key information for the customer toward their objective of developing USP and DSP processes that are amenable for full scale manufacture.
Here at Sygnature Discovery we support many aspects of biopharmaceutical development. Not just elucidation of CQAs with mass spectrometry and other techniques but also experiments to refine USP and particularly DSP at lab scale aiming to develop processes that are robust, reproducible and therefore amenable for full scale manufacture. If you think your project would benefit from our input, then please don’t hesitate to get in touch info@peakproteins.com with us.
References
[1] A. Eon-Duval, H. Broly, and R. Gleixner, “Quality attributes of recombinant therapeutic proteins: An assessment of impact on safety and efficacy as part of a quality by design development approach,” Biotechnol Prog, vol. 28, no. 3, pp. 608–622, May 2012, doi: 10.1002/btpr.1548.
[2] A. S. Rathore and H. Winkle, “Quality by design for biopharmaceuticals,” Nat Biotechnol, vol. 27, no. 1, pp. 26–34, Jan. 2009, doi: 10.1038/nbt0109-26.


