AlphaFold for fast structural determination of a previously unsolved domain
AlphaFold
AlphaFold is a deep learning system, developed by DeepMind that predicts a protein’s 3D structure from its amino acid sequence. After publication in Nature in 2021, AlphaFold was subsequently used across the entire human proteome. This has since expanded to 48 organisms date which are all accessible using the AlphaFoldDB database. AlphaFold has become a valuable tool for Drug Discovery. At Peak Proteins we regularly use AlphaFold as an aid in construct design. Another great example is using these structural models to help structural biologist model experimental structural data.
Using AlphaFold as a model for molecular replacement
We have been working on a multi-domain integrin protein which exhibits large conformational movements. One of the challenges for the structural determination of this protein was a previously unsolved and flexible terminal domain.
There are several ways to model a previously unsolved domain. Mainchain tracing can be used to sequentially add amino acids into unmodelled electron density. However, in our case poor electron density and several disulphide bonds made mainchain tracing difficult. Homology modelling is another commonly used method where a different protein structure with a similar amino acid sequence is used as an initial model for molecular replacement. In our case there was a homology model available for the domain but attempts to use it for molecular replacement were unsuccessful (Figure 1b). The AlphaFold model of the domain was used as an alternative search model in molecular replacement. This model did provide us with a solution, and it was impressively close to the final refined structure of the domain (Figure 1a).
This provides another example where AlphaFold has provided a very good initial model for structural determination and saved a significant amount of time compared to other methods.
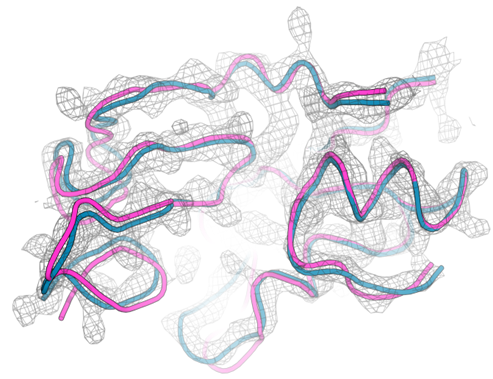
Figure 1: Using AlphaFold as a modelling tool. a) AlphaFold model (pink) superimposed onto final refined model (blue) with electron density map shown in gray. b) homology model (yellow) superimposed onto final refined model (blue) with electron density map shown in gray.
If you would like more information about our structural biology services at Peak Proteins, please contact us.


