Protein Expression – A signal Something is Wrong
A few months ago, we had a succession of proteins we thought should be well expressed following secretion from the HEK293-6E cell system we use. However, expression was significantly below what we expected making it difficult to supply protein for biophysical or crystallographic studies. We wondered if the HEK cells had reached too high a passage number or if there was a problem with the cell culture medium but a positive expression control protein construct, we always use looked fine. Perhaps plasmid quality was poor so we remade the positive control DNA alongside the misbehaving constructs. The positive control again worked fine but the mis-behaving constructs didn’t.
We then came across a paper by Gülin Güler-Gane et. al., 2016 that described the influence of different amino acids at the +1 position after the signal peptide showing that certain amino acid residues cysteine (C), proline (P), tyrosine (Y) and glutamine (Q) can be detrimental, and that inserting an alanine can sometimes fix these problems. Some but not all of our constructs did have a Q at position +1.
Two other observations occurred to us. Firstly, we often, but not always, use a generic mammalian signal peptide of Ig kappa (Igk) and wondered if the native signal peptide might be preferred. Secondly, we knew from literature precedent that one construct starting signal peptide-6His-protein should be successful whereas our construct was IgK-6His-Avi-TEV-protein was not. We wondered if the presence of the TEV and/or Avitag might be the issue. Addressing these issues yielded mixed results with no obvious fix-all solution. Below are project examples where we tried to address expression of the different secreted proteins.
Additional alanines after the signal sequence did not help expression
Out of 4 large viral glycoproteins we expressed, two proteins from the betacoronavirus family expressed well and two from the alphacoronavirus family did not express well, with one in particular giving very poor yields; this particular construct had a Q at +1. We tested out the +1 position hypothesis and inserted either 1 or 2 alanines after the Ig K signal peptide and additionally, we tested the native signal peptide. No improvement in the yield of expressed protein was seen, suggesting that the poor expression was not due to the signal peptide per se but some unknown structural or biological function of the protein that may limit its recombinant expression.
Changing to the native signal peptide can make a difference.
We expressed a chemokine where we initially use our standard Igk signal peptide but surprisingly obtained poor expression. The signal peptide was then switched to the native signal peptide; this improved expression ~ 10-fold compared to Igk.
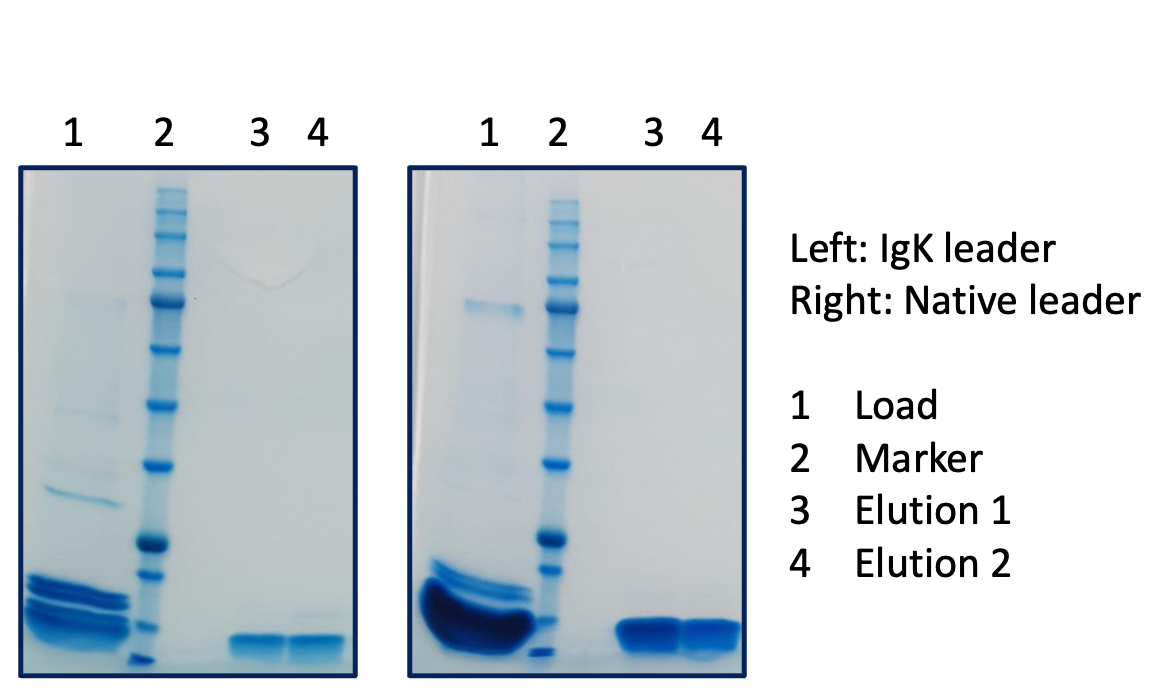
In a second example of a secreted receptor, we compared the native and Igk signal peptides with 1 or 2 Ala inserted. In this case the native signal peptide gave ~3-fold improvement in expression but inserting alanine again gave no additional improvements.
Additional tag sequences can dramatically reduce expression of a secreted protein.
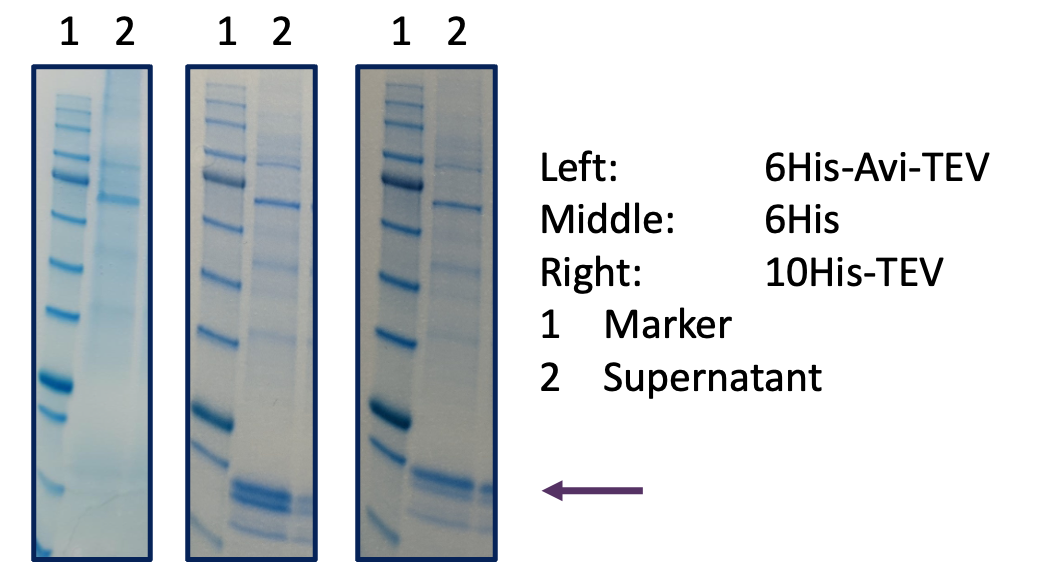
In this example, we wondered whether the insertion of a TEV and/or Avi sequence at the N-terminus after the Igk was causing expression problems. In two separate experiments, the 6His-Avi-TEV construct expressed at very poor levels of ~50 µg/L whereas a positive control protein expressed well. While still retaining the Igk, we examined N-terminal 10His-TEV and 6His tags. Both these new constructs expressed at levels approaching 50 mg/L, almost 3 orders of magnitude better than the original construct, suggesting the Avi tag was the problem. 10His immobilisation for SPR studies was successfully used by the client as an alternative to Avi-tagged biotinylated protein.
So what did we learn?
Unsurprisingly, with working with proteins, our conclusions are mixed. In agreement with by Gülin Güler-Gane et. al., 2016 we see that both the signal peptide and mature protein sequence are important for recombinant expression. Inserting alanine did not work for us with these proteins but we did see a detrimental effect in one case from the mature protein sequence through using additional tag sequences. We did observe dramatic improvements in some cases however from changing our standard signal peptide to the native sequence. This indicates that in some cases the natural protein sequence is preferential to our engineered IgK sequence which is typically used because of its high secretion properties.
These results further emphasise that proteins are very much individual molecules and recombinant protein production has to be tailored to each protein. Here at Peak Proteins, we have a wealth of experience in expressing and purifying proteins and treat each bespoke protein we make with a research-based approach, as exemplified above, to ensure protein are produced to meet our clients’ project needs.


