The Naming of Cats (Proteins)
With apologies to T. S. Eliot
The Naming of Proteins is a difficult matter,
It isn’t just one of your holiday games;
You may think at first I’m as mad as a hatter
When I tell you, a protein must have THREE DIFFERENT NAMES
First of all, there’s the name that the family use daily…..
This is the abbreviation banded around the lab and office to refer to the protein in question, such as YAK1. This is the common-or-garden name “Yet Another Kinase 1” that you should be able to link straight to the Uniprot database entry. Quoting the Uniprot reference defines which species the protein comes from, but of course there is still possible ambiguity at this level. The common name could well refer to more than one form of the protein; isoforms or sequence variants, engineered constructs, activated versus precursor forms or mutant versions perhaps.
For these common protein names there are International guidelines for protein naming and nomenclature that have been produced by the European Bioinformatics Institute (EMBL-EBI), the National Center for Biotechnology Information (NCBI), the Protein Information Resource (PIR) and the Swiss Institute for Bioinformatics (SIB). Their intention is to promote consistency in protein naming across databases, which will in turn aid data retrieval and improve communication.
Some of the more quirky names given to proteins have wonderful stories behind them and a few of our favourites follow.

Sonic was named because a research team decided to name each of the related proteins that they had discovered after hedgehogs. Robert Riddle a postdoc fellow was heavily into a band called Sonic Youth and by coincidence happened to see an ad for the new at the time Sonic the Hedgehog computer game.

The popular children’s dolls Ken and Barbie lent their name to a protein because they are apparently sexless. The protein now with that name can result in sterility in fruit flies whereby both male and female fruit flies have no external sex organs.

Mutations in Spock1 results in Zebrafish with pointy ears.
JAK1 and 2 were originally known as Just Another Kinase. However,  because they have two almost identical domains they were renamed as Janus kinase 1 and 2 after the two-faced Roman god of beginnings, endings, transitions and weirdly doorways.
because they have two almost identical domains they were renamed as Janus kinase 1 and 2 after the two-faced Roman god of beginnings, endings, transitions and weirdly doorways.
But I tell you, a protein needs a name that’s particular,
A name that’s peculiar, and more dignified,…..
Now we are into the realms of recombinant constructs where the detail of the particular form of the protein is described and defined. A singular, specific, unambiguous….
….. Names that never belong to more than one protein.
Whilst there are common nomenclature conventions for genes [1] [2] [3] [4] and for proteins themselves [5] the naming of recombinant protein constructs in the scientific literature is exceptionally diverse with a variety of approaches displayed by researchers. Even within our own small organisation there are differences in the way people write out the names of their protein constructs.
At this level to remove any ambiguity there is a need for a system to define both the species that the protein of interest derives from along with the exact form and composition of the protein construct. When discussing a protein, where possible the Uniprot [6] reference should be quoted for the reasons stated earlier.
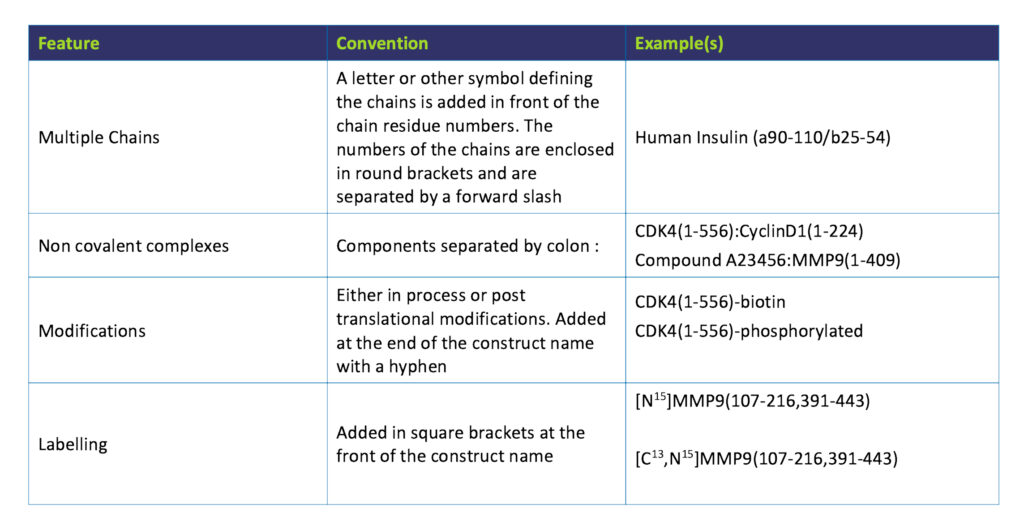
Furthermore, we need to be able to differentiate between what was originally cloned and the protein construct that is finally purified as they could very well be different after the removal of tags for example. We also need to be able to take into account any modifications (in process or post translational) that may have occurred along the way. In our labs we have a set of guidelines (rather than rules) that are intended to make protein construct naming as consistent as possible. A table summarising this convention is given below and we hope it may be of use to you too.
But above and beyond there’s still one name left over,
And that is the name that you never will guess;
The name that no human research can discover—
But THE PROTEIN HERSELF KNOWS, and will never confess.
When you notice a protein in profound meditation,
The reason, I tell you, is always the same:
Her mind is engaged in a rapt contemplation
Of the thought, of the thought, of the thought of her name:
Her ineffable effable
Effanineffable
Deep and inscrutable singular name.
Peak Proteins’ Protein Construct Naming Convention
Compile the various features of the protein construct using the guidelines given in the table below and then assemble them in the order that they occur in the protein sequence. For complete unambiguity the Uniprot [6] reference number should be quoted and ideally the actual amino acid sequence of the construct should be included in the paper or report.

Example: We have cloned a human matrix metallo proteinase 9 (uniport P14780) construct with just the pro-peptide and the main chain preceded by a 6His tag that has a TEV cleavage site after it. The construct is to be transfected into HEK293 cells so we have also added a signal sequence from honey bee melittin to promote secretion into the media during culture. The cloned construct name would be:-
Apis mel. Melittin sig. (1-21)-6His-TEV-Pro(20 – 93)-MMP9(94 – 707)
Or if we didn’t need to pass on detail about the signal sequence and the pro domain we could just use this simpler construct name which would also be correct:-
6His-TEV-MMP9(20 – 707)
During purification we cleave the tag off using TEV and perform an autoactivation step that releases the mature active MMP9. Because we expressed it in HEK cells we have managed to achieve glycosylation at positions 120 and 127. Therefore, the final purified protein name would be:-
MMP9(107 – 707)-[N glyco Asn120, Asn127]
References
[1] HUGO Gene Nomenclature Committee, “The resource for approved human gene nomenclature,” [Online]. Available: https://www.genenames.org/.
[2] den Dunnen J T et al, “HGVS Recommendations for the Description of Sequence Variants: 2016 Update,” Human Mutation, vol. 37, no. 6, pp. 564 – 569, 2016.
[3] Human Genome Variation Society, “Sequence Variant Nomenclature,” [Online]. Available: http://varnomen.hgvs.org/.
[4] Wikipedia, “Gene Nomenclature,” [Online]. Available: https://en.wikipedia.org/wiki/Gene_nomenclature.
[5] NCBI, “International Protein Nomenclature Guidelines,” [Online]. Available: https://www.ncbi.nlm.nih.gov/genome/doc/internatprot_nomenguide/.
[6] UniProt Consortium, “UniProt,” [Online]. Available: https://www.uniprot.org/.


