Development of a Refolding Protocol for an Interleukin
One of our Senior Protein Scientists, Sam Bloor, describes the journey to develop a refolding route for an interleukin.
Introduction
Cytokines are important cell signalling proteins which bind to cell surface receptors, initiating downstream cascades 1. They have a range of functions such as autocrine and endocrine signalling, as well as modulating our immune systems. One class of cytokines, known as interleukins, have been of particular pharmacological interest due to their role in the immune system1. Interleukins are expressed and then secreted by white blood cells2.
Peak Proteins (Part of Sygnature Discovery) have expressed, purified, and characterised a variety of interleukins for numerous clients. These proteins have been expressed in a range of host systems such as E. coli, insect and mammalian cells, so that post translational modifications could be tailored for the specific end use of the client. For instance, a method to refold an interleukin (IL-X) from inclusion bodies produced in E. coli was developed so that the protein could be used in structural studies (crystallography/NMR). The same IL-X was also expressed in parallel using HEK293 cells for assay purposes. Expression by HEK293 cells resulted in a heavily glycosylated IL-X protein which would have been incompatible with structural studies but provided a more native protein for biochemical characterisation. We can deglycosylate a protein using a combination of kifunensine and EndoH but in this case NMR was also being considered and E. coli expression would also allow for isotopic labelling.
The Steps to Develop a Refolding Protocol
Whilst expression and purification of IL-X from HEK293 cells was relatively straightforward, the refolding of IL-X from inclusion bodies required a preliminary method development study to optimise the process. Targeting recombinant protein expression to inclusion bodies rather than soluble expression is a useful tool to obtain insoluble proteins for subsequent solubilisation and refolding. Inclusion bodies are typically formed by expression of a protein at 30 oC for 4-6 hours following induction with 0.1 – 1.0 mM IPTG; however, we followed the literature precedent for IL-X and found that for optimal yield the cells should be incubated for a further 18 hours at 30 – 37 oC following induction with IPTG.
Given that during our inclusion body preparation a considerable number of impurities remained (Figure 1), we decided to include a denatured immobilised metal affinity chromatography (IMAC) purification step. Denatured IMAC follows the same principles as soluble IMAC in that an affinity tag (6His-, 10His-) binds to a metal ion (Ni2+, Co2+) that is chelated on the resin beads; however, denatured IMAC is performed in the presence of a chaotrope such as urea or guanidine to keep the protein in a soluble unfolded state. In this instance IL-X was the only protein bound to the denatured IMAC column and eluted as expected using an imidazole gradient (Figure 2). The impurities which lacked an affinity tag (or intrinsic metal affinity) did not bind to the resin and therefore passed through the column.
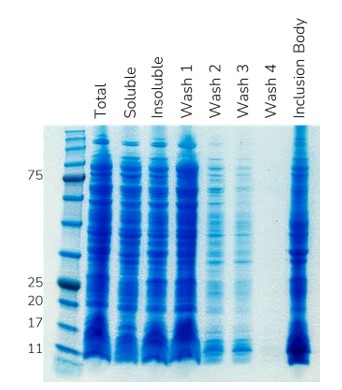
Figure 1. SDS-PAGE image showing inclusion body preparation of IL-X. A considerable amount of impurities remain in the inclusions bodies despite multiple rounds of washing.
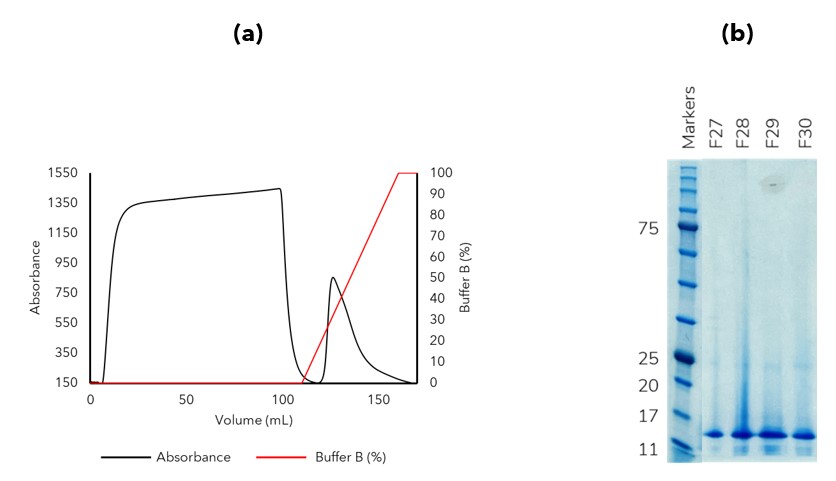
Figure 2. Denatured IMAC was used to purify the IL-X protein prior to refolding. (a) AKTA trace (b) SDS-PAGE image of elution fractions (some lanes omitted from image for clarity)
Following denatured IMAC, we were able to obtain an accurate estimation of the protein concentration and could therefore tailor the refolding process accordingly. IL-X was then refolded by rapid dilution to reduce the chaotrope and imidazole concentration, followed by sequential dialysis steps to reduce the salt concentration.
Capture of the refolded IL-X protein by ion exchange chromatography (IEX), required multiple development steps where we evaluated various factors including refolding buffer components to refine the protocol. Initial attempts at IL-X capture contained too high a salt concentration to be compatible with IEX (Figure 3a), however, diluting the sample 1:1 in low salt buffer to further reduce the salt concentration and conductivity after dialysis resulted in an 12-fold increase in IL-X capture (Figure 3b). The protein was then purified by size exclusion chromatography (SEC) as a final polishing purification step to remove any soluble aggregates from the sample.
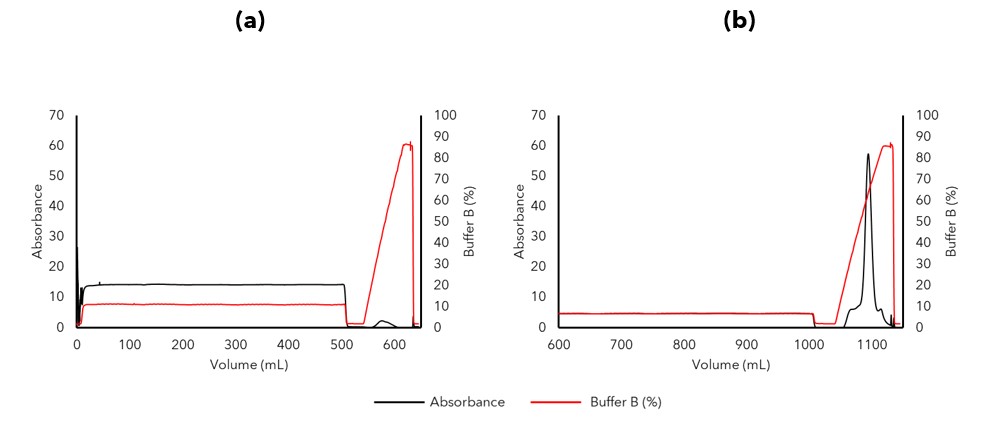
Figure 3. IL-X was captured using an ion exchange chromatography column. (a) Initial attempts at IL-X capture resulted in a low yield due to inadequate salt reduction by dialysis. (b) Dilution of the post dialysis refold 1:1 in lower salt buffer reduced the salt concentration further and the buffer system was then compatible with increased capture by IEX.
The Impact
Following this study a post translational modification free, refolded E. coli IL-X protein was successfully produced to support structural studies (Figure 4a), in parallel with a post translationally modified (glycosylated) IL-X from HEK293 cell expression for biochemical characterisation (Figure 4b). The E. coli produced protein was shown to be similarly folded to the IL-X protein produced by HEK293 cells by 1D NMR.
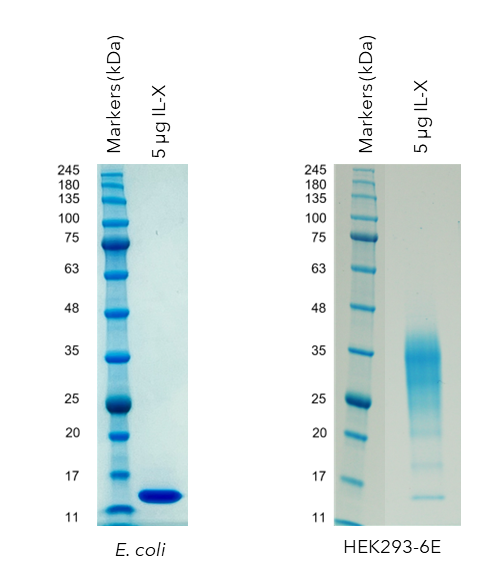
Figure 4. SDS-PAGE analysis of IL-X produced in either (a) E. coli or (b) HEK293 cells. The IL-X protein produced by E. coli is a single, well defined, band corresponding to an unmodified IL-X protein. One the other hand, the IL-X protein produced in HEK293 cells displays an expected smear pattern which is indicative of glycosylation post translational modifications.
Your Refolding Project
The team at Peak Proteins have developed refolding protocols for a wide range of protein targets including several cytokines, such as the example described above in this case study. If your project would benefit from our expertise in this area then please don’t hesitate to get in touch. We would be more than happy to discuss your particular needs. info@peakproteins.com
References
1. Propper, D. J. & Balkwill, F. R. Harnessing cytokines and chemokines for cancer therapy. Nature Reviews: Clinical Oncology. 19. 237-253. (2022)
2. Briukhovetska, D. et al. Interleukins in cancer: from biology to therapy. Nature Reviews: Cancer. 21. 481-499. (2021)

