Characterising Intrinsically Disordered Proteins
In this case study Toby Allen demonstrates the value of using multiple analytical methods to characterise Intrinsically Disordered Proteins (IDPs) and highlights some of the techniques we have available at Sygnature Discovery.
What are IDPs:
While many globular proteins have been characterised with a stable tertiary structure, Intrinsically Disordered Proteins (IDPs) are characterised by multiple, unstable, and predominantly local structures. Many proteins exist on a spectrum between these extremes, as IDPs can contain structured regions, or structure can be induced following ligand binding or protein-protein interactions. For example, many transcription factors are IDPs where structure is induced on binding of their target DNA. Often IDPs have no single well-defined equilibrium structure and exist as highly dynamic, heterogeneous ensembles of conformers [1]. There is growing interest in IDPs as a class of proteins, with citations relating to IDPs increasing exponentially since 1995.
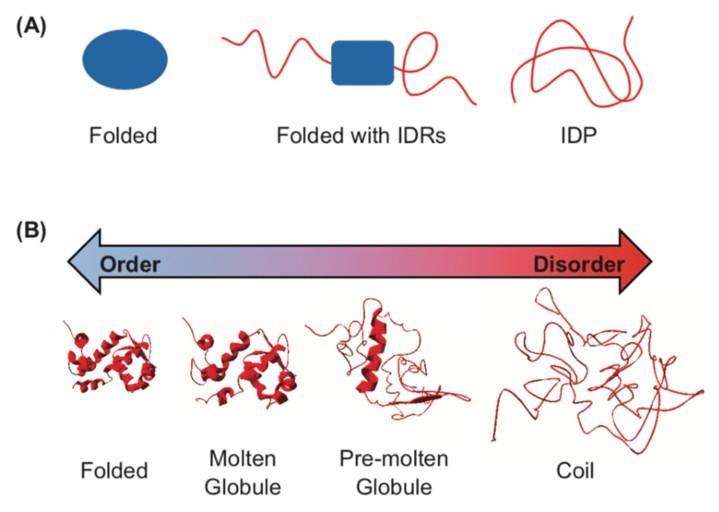
Figure 1: the spectrum of protein structure – between folded and random coil (IDP) [2].
Project Background
Recently within the Protein Science department at Sygnature Discovery we have been working on one of these IDPs – a transcription factor expressed as insoluble inclusion bodies in E. coli. We purified this protein target by isolating the inclusion bodies followed by solubilisation and refolding. The protein was well expressed and purified to homogeneity by SDS-PAGE – see figure 2:
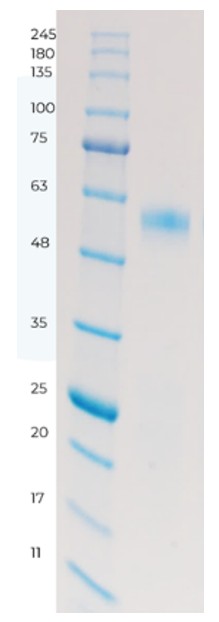
Figure 2: SDS-PAGE showing the target is >95 % pure
During a standard analytical SEC run we noticed the protein eluted in the void volume of the column – suggesting it is far larger than we would expect from the molecular weight. This is consistent with our target as an IDP, as the lack of global structure can cause IDPs to be less compact and therefore to run larger than expected.
Unfortunately, this can limit the usefulness of the analysis as we can’t easily distinguish monomeric protein from oligomers or high-molecular weight aggregates when they elute in the void. The protein was working in the assays, but our assay team were seeing some unusual results that might indicate the protein had formed a dimer.
Alphafold is useful for predicting the structure of proteins from the sequence. In this case the structure of the target is predicted to be largely disordered – but with a leucine zipper (LZ). Leucine zippers are known to form dimers – this may be giving us another clue about what state our target may be in.
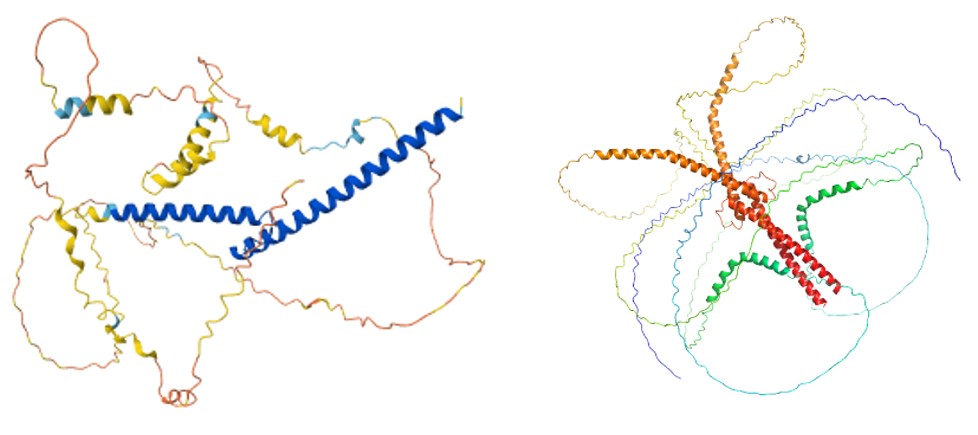
Figure 3: AlphaFold predicted models of 1 x IDP and 2 x IDP, showing predicted dimerisation
To ensure we were providing our client with the high-quality protein they needed, we used a range of the characterisation techniques we have available to build a comprehensive picture of the target.
SEC-MALS:
SEC-MALS (Size Exclusion Chromatography with Multi-Angle Light Scattering) is something of a ‘gold standard’ for protein characterisation and is particularly useful for IDPs as it allows separate measurement of the protein size and weight. Proteins are first separated using size exclusion then the molecular weight of eluted species is determined using light scattering. Unfortunately, in this case we weren’t able to complete this analysis as the protein did not elute from the high molecular weight columns, and characterising potential multiple species eluting in the void volume of the standard column wouldn’t have been definitive. SEC-MALS methods are often protein dependent so this is not surprising, and it may be possible to get better data with further buffer optimisation. For now, we decided to use other QC techniques to characterise the target.
Differential Scanning Fluorimetry (DSF):
In DSF a hydrophobic fluorescent dye is incubated with the protein of interest, but in solution water quenches the fluorescence of the dye. The mixture is heated and changes in fluorescence are monitored as temperature increases. The dye binds to the hydrophobic core of the protein as it unfolds – excluding the water and therefore increasing the fluorescence.
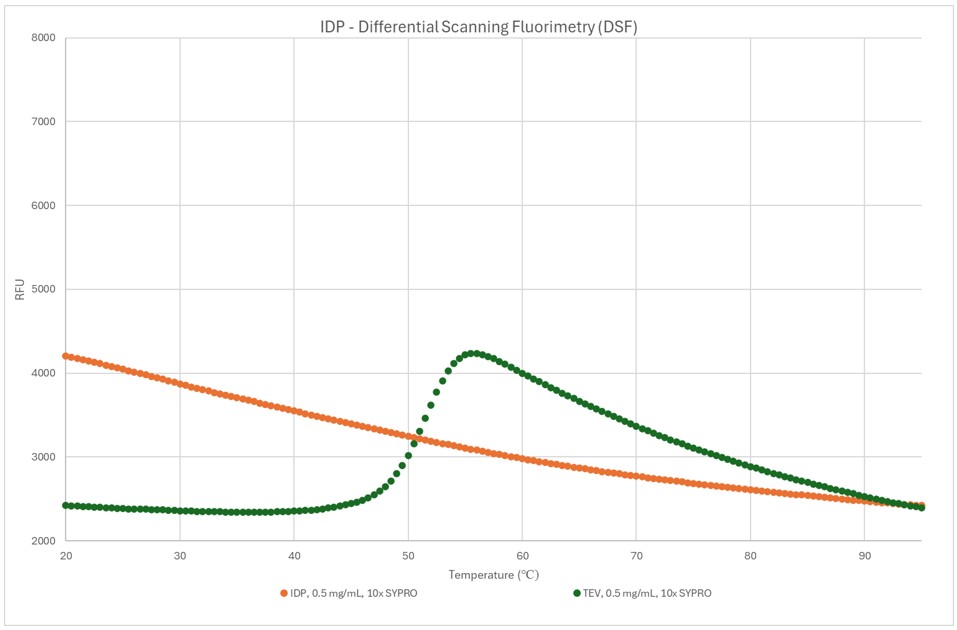
Figure 4: DSF trace showing IDP target vs TEV protease control
This technique is well-established for globular soluble proteins, and TEV protease was used as a control in this assay as it is known to be fully folded and relatively stable. It shows the characteristic increase in fluorescence as temperature approaches 50 degrees as the protein begins to fully unfold.
The orange trace shows that the IDP we are trying to characterise has no clear unfolding transition. This is characteristic of an intrinsically disordered protein as denaturing doesn’t expose a hydrophobic core. At first glance, the high initial signal might suggest the protein is aggregated, but as we could reproduce this curve through multiple rounds of heating and cooling we believe it is an artifact, and other techniques would be more useful for this target.
Circular Dichroism:
Circular Dichroism (CD) is a useful technique for characterising secondary structure in proteins by examining the difference in absorption of left-handed and right-handed circularly polarised light.
Different secondary structural elements have different characteristic CD spectra – Alpha-helixes have characteristic minima at 222 and 208 nm, B-sheets at 218 nm, and disordered proteins (random coils) have low ellipticity above 210 nm and a minimum near 195 nm:
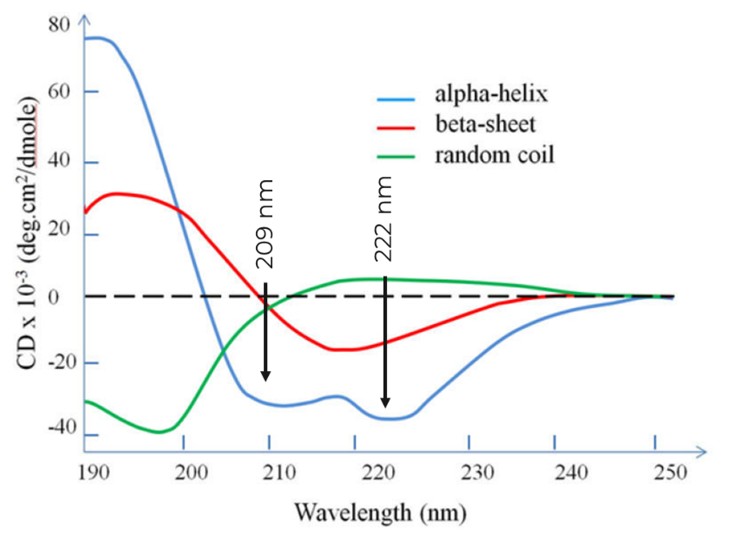
Figure 5: characteristic CD spectra of different elements of secondary structure
Given the predicted structure of the target being predominantly disordered with some alpha helical structure, we collected a full UV CD spectrum from 180-260 nm. We collected three data sets – initially at 5oC , then after heating the sample to 80oC and finally after cooling back down to 5oC.
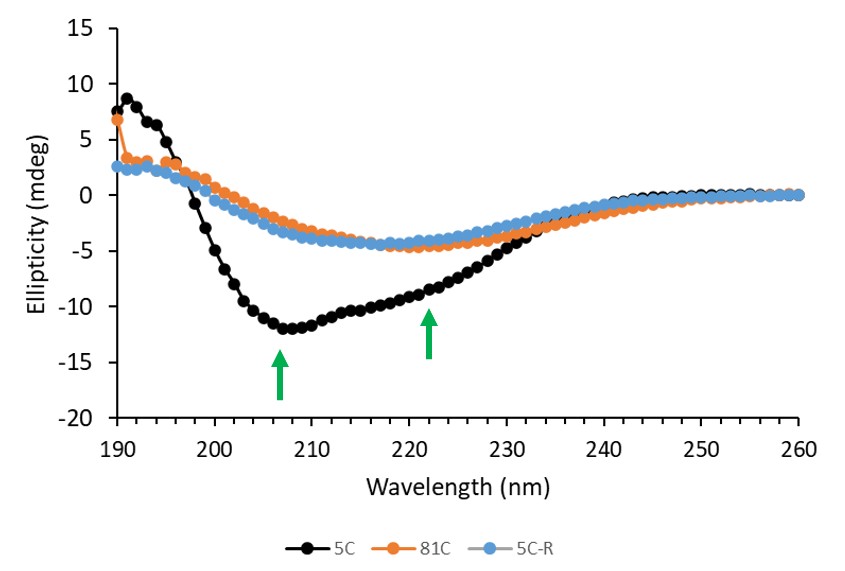
Figure 6: CD spectra of IDP target at 5, 81, 5 degrees centigrade
In the initial measurement, the target displays a clear minimum at 208 nm, and to a lesser extent 222 nm. This is characteristic of alpha helices but seems to be distorted by a significant contribution from a random coil. This backs up the prediction of a predominantly disordered protein with some alpha-helical character from the leucine zipper. The minima disappear when the sample is heated, implying a loss of these structural elements. Together, this indicates that the protein does initially contain some structured regions, and that these are irreversibly denatured upon heating.
Melting temperature of the target was also determined, around 62oC. A single denaturation transition was observed around 55-69oC- see below. This provides evidence of some elements of structure in the target.
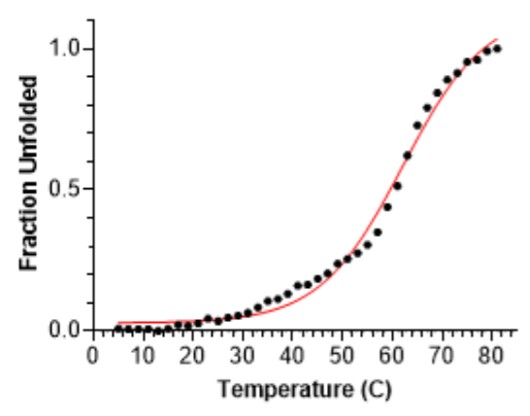
Figure 7: unfolding curve of IDP target
The CD analysis suggested that the protein we prepared and supplied contained the correct structural features.
FIDA:
FIDA allows the measurement of hydrodynamic radius (Rh) in solution by examining how the sample diffuses under laminar flow conditions. (See our FIDA blog here for an overview). We can use FIDA to analyse unlabelled proteins using their intrinsic tryptophan fluorescence, and the measured hydrodynamic radius (Rh) values can be compared to reference values calculated from PDB structures or to theoretical values for proteins of a similar molecular weight.
We analysed our protein using FIDA and compared the measured size to values predicted for monomers and dimers as both perfect globule and linear peptides.
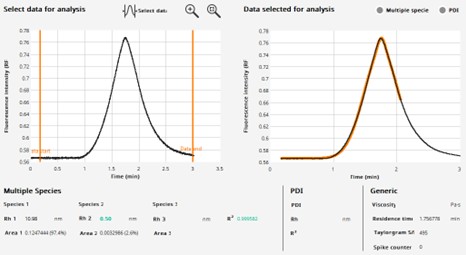
Figure 8: FIDA spectra of IDP target
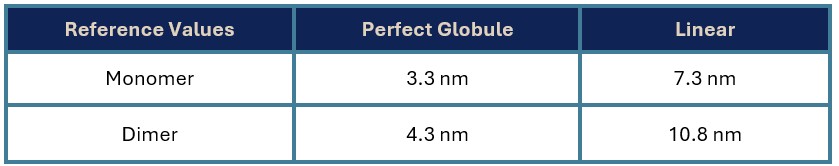
Table 1: hydrodynamic radius reference values for the target
The measured Rh (10.98 nm) is closest to the predicted value for a linear dimer – and this is the first evidence that the protein is likely to be dimeric as suspected and predicted by Alphafold. The value being close to the predicted linear size is consistent with a predominantly disordered protein.
When we analysed the protein in the presence of 5M urea, we observed that the measured size decreased to 6.2 nm – consistent with the predicted size of the linear monomer and indicating that we have disrupted the dimeric species.
Critically, we do not observe any very large species indicating aggregates – these would appear as spikes in the un-processed trace on the left.
Together, the FIDA analysis suggest the target is a disordered dimer in solution.
Summary:
Given that SEC-MALS was not able to provide us with a molecular weight, characterising this target initially provided challenging. By combining important data using a range of techniques available to us, we have assembled evidence that the target has been produced as a soluble, predominantly disordered, dimeric species – as predicted by Alphafold. We were confident that we could use this protein for our client’s ongoing assay development.
For all your protein needs – whether soluble, disordered, or membrane proteins please get in contact info@peakproteins.com
References:
1. Uversky V N, Introduction to Intrinsically Disordered Proteins (IDPs). Chemical Reviews. (2014), 114 (13), 6557-6560, DOI: 10.1021/cr500288y
2. Watson M, Stott K, Disordered domains in chromatin-binding proteins. Essays Biochem. (2019), DOI: 10.1042/EBC20180068


