Determining the Cryo-EM Structure of the 5-HT2A receptor with Serotonin Bound
In this case study, Rachel Johnson describes how we determined the active state structure of the 5-HT2A receptor with the natural ligand serotonin bound using cryo-EM.
Introduction
GPCRs are a large and diverse group of receptors in the body. They are also important therapeutic targets as over a third of all FDA approved drugs act upon these receptors [1]. Being able to probe how ligands interact with the target protein by elucidating the structural details of how the ligand binds to the protein and/or revealing any changes to the conformation of the protein can significantly impact drug discovery programs. Cryo-EM has become a powerful tool in the structure determination of GPCRs [2]. The resolutions attained have allowed small molecule ligands to be visualised within the maps and have also revealed different receptor conformations upon ligand binding and enabled receptor dynamics to be studied [3, 4].
The 5-hydroxytryptamine receptor isoform 2A (5-HT2AR) is one of a family of GPCRs activated by serotonin (5-hydroxytryptamine) and is essential for learning and cognition [5]. Pathophysiology associated with 5-HT2AR has been linked to psychiatric disorders and conditions including depression, schizophrenia, and drug addiction. In addition to serotonin, 5-HT2AR is also the target for agonists that possess cognition-enhancing and hallucinogenic properties, such as LSD and psilocin. On the other hand, 5-HT2AR antagonists have antipsychotic and antidepressant properties. Thus, 5-HT2AR has seen recent focus as a possible target in the treatment of a range of neuropsychiatric conditions [6]. Here, we aimed to determine the active state structure of 5-HT2AR with the natural ligand serotonin bound using cryo-EM.
Sample preparation
Initially, we co-expressed the receptor and G proteins in Sf21 insect cells using the baculovirus expression system where the components of the complex (5-HT2AR, Gαq, Gβ1 & Gg2) were added at a 1:1:1:1 ratio. Following an incubation with serotonin, the complex was purified from membranes via FLAG affinity and size exclusion chromatography (SEC). Sample quality is crucial for cryo-EM, so final QC checks were performed. SDS-PAGE and MS-MS peptide mapping was used to ensure all complex components were present. An analytical SEC was also performed to check that the protein could withstand a freeze-thaw cycle. This is important because if complexes are unstable during freeze-thaw, they must be applied to a cryo-EM grid immediately after purification for optimal structure determination. QC checks for5-HT2AR confirmed all components of the complex were present in the final sample and that the complex survived a freeze-thaw cycle. Therefore, the sample was taken forward for imaging, and frozen sample could be used.
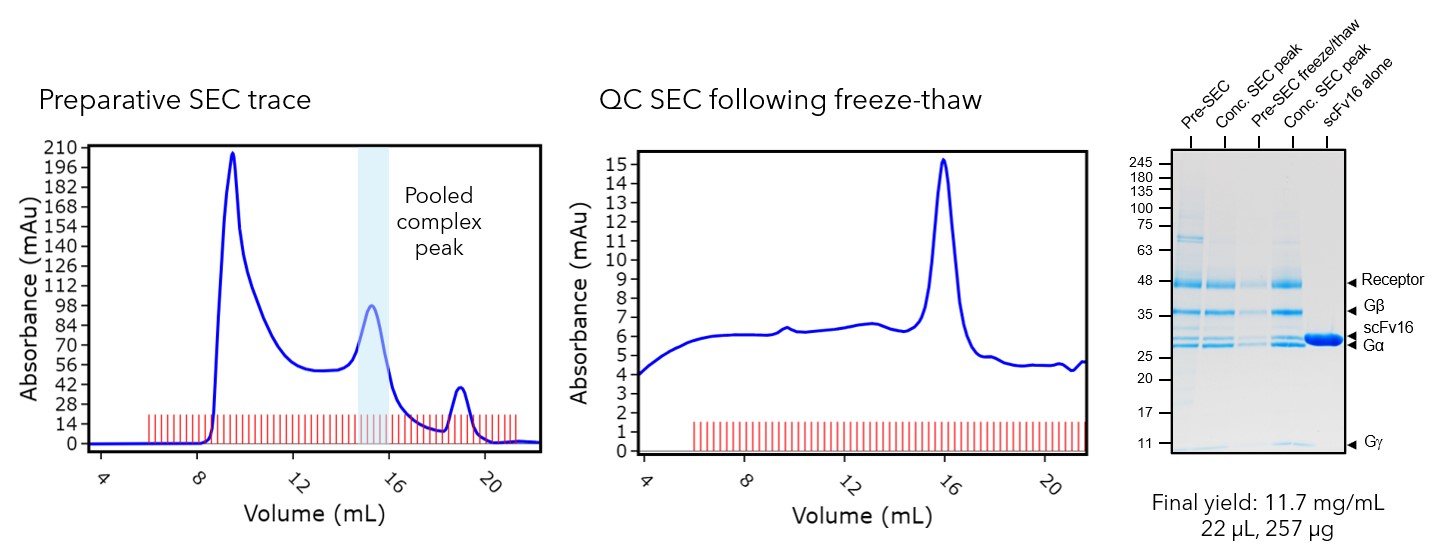
Figure 1: Protein purification and quality control of the 5-HT2AR sample for cryo-EM. Fractions from the preparative SEC trace peak were pooled. The freeze-thaw and SDS-PAGE QC checks showed that the all components of the complex were present and that it could withstand a freeze-thaw cycle.
Cryo-EM structure determination
Cryo-EM grids of the 5-HT2AR complex were prepared using a ThermoFisher Vitrobot Mark IV. 3 µL of complex was applied to a Quantifoil R1.2/1.3 Cu 400 mesh grid, excess sample was blotted off and the grid was plunged into liquid ethane rapidly freezing the sample in a thin layer of vitrified ice to preserve its native state. Prepared grids were screened for good particle distribution in thin ice. Suitable regions of the grid were identified and a data set consisting of ~11,000 movies was collected on the Titan Krios microscope fitted with a K3 detector at the Midlands Regional Cryo-EM Facility. After motion correction and CTF refinement, particles were picked using Topaz [7] and classified in 2D using Relion5 [8]. The particles which contributed to good 2D averages that displayed clear secondary structure were then used for 3D refinement and subjected to particle polishing which resulted in a 3.7Å map being obtained. Atomic coordinates were then fitted into the polished map and subjected to real space refinement.
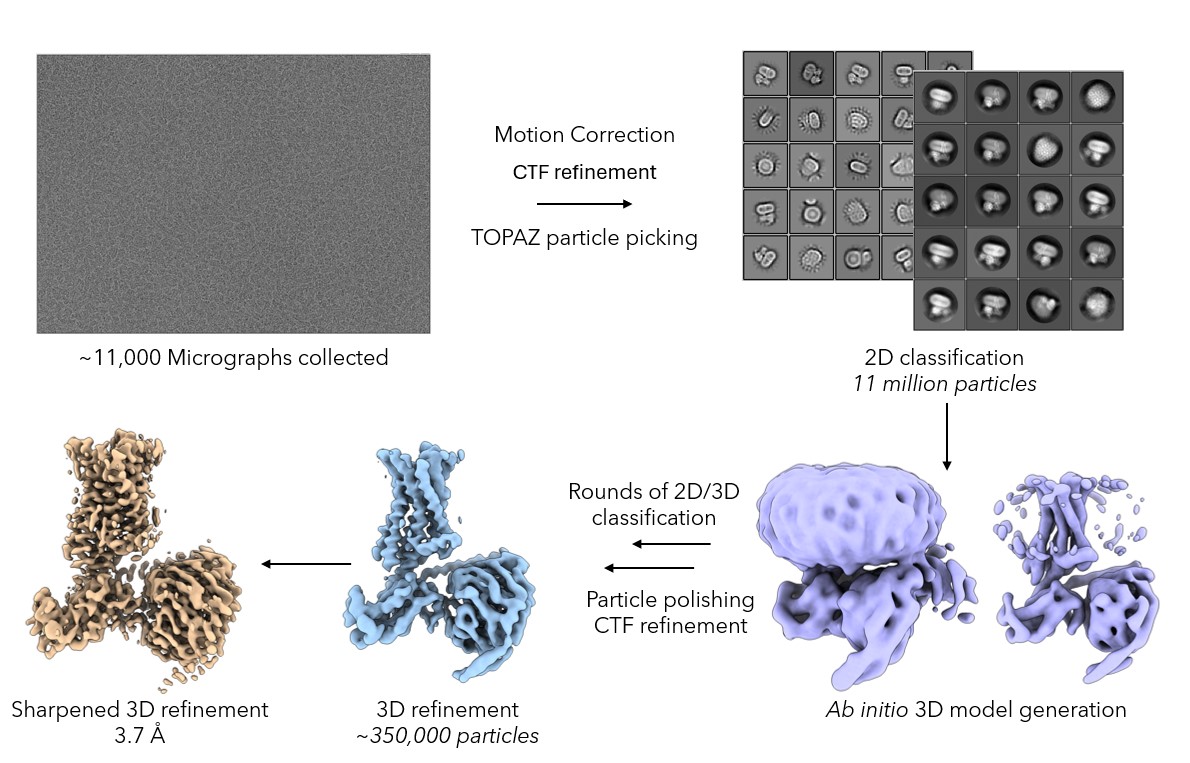
Figure 2: The cryo-EM data processing pipeline. Particles were picked from ~11,000 movies using TOPAZ and a 3D initial model was generated that showed secondary structure features. Particles were subjected to iterative rounds of 2D and 3D classification to generate a homogenous particle stack. These particles underwent 3D refinement, particle polishing and CTF refinement before a 3.7Å map was obtained.
Cryo-EM structure analysis
The 5-HT2AR map quality allowed the receptor and G protein components to be modelled into the map. Analysis of the ligand binding site also showed strong density for serotonin and the surrounding receptor side chains allowing the compound to be unambiguously modelled into the map. We compared our cryo-EM structure to two previously published structures. The first is a cryo-EM structure of the active state 5-HT2AR with 25CN-NBOH bound (PDB: 6WHA) and the second is a receptor-only X-ray crystallography structure with serotonin bound (PDB 7WC4). Interestingly, the serotonin binding pose in our cryo-EM structure agrees with the ligand binding site in the 25CN-NBOH small molecule agonist cryo-EM structure. This differs from that in the X-ray crystallography structure, suggesting that relying solely on X-ray data may be misleading to small-molecule medicinal chemistry efforts in this case.

Figure 3: 5-HT2AR cryo-EM structure analysis. A) The 5-HT2AR map coloured by complex component. The ligand binding site is highlighted by the black box. B) Analysis of the ligand binding site showed strong density for serotonin and the neighbouring side chain residues. C) An overlay of our cryo-EM structure (green) with the 25CN-NBOH (purple) cryo-EM structure and the X-ray crystallography structure with serotonin bound (grey). Serotonin binds at the same site as 25CN-NBOH which differs from the serotonin bound X-ray structure.
Conclusions
We determined a cryo-EM structure of 5-HT2AR where the resolution attained was sufficient to unambiguously model the small molecule ligand into the map. Here at Sygnature Discovery (Peak Proteins), we have the capability and expertise to determine cryo-EM structures of GPCRs. We can produce high quality protein samples, prepare cryo-EM grids in-house with fresh protein and access cryo-EM facilities across the UK allowing us to obtain high resolution structures. For more information on how we can use cryo-EM to support your GPCR drug discovery programs, please contact info@peakproteins.com
References
1. Sriram, K.; Insel, P. A.; G Protein-Coupled Receptors as Targets for Approved Drugs: How Many Targets and How Many Drugs? Mol Pharmacol. 2018 93 (4), 251-258.
2. Zhang, X.; Johnson, R. M.; Drulyte, I.; et al. Evolving cryo-EM structural approaches for GPCR drug discovery. Structure. 2021, 29, (9), 963-974.e6.
3. Cao, J.; Belousoff, M. J.; Liang, Y. L.; Johnson, R. M.; et al. A structural basis for amylin receptor phenotype. Science. 2022 Mar 25;375(6587):eabm9609.
4. Zhang, X.; Belousoff, M. J.; Zhao, P.; et al. Binding and Activation by Peptide and Non-peptide Agonists. Mol Cell. 2020, 80, 485-500.
5. Zhang, G.; Stackman, R. W. The Role of Serotonin 5-HT2A Receptors in Memory and Cognition. Front. Pharmacol. 2015, 6.
6. Cameron, L. P.; Benetatos, J.; Lewis, V.; et al. Beyond the 5-HT 2A Receptor: Classic and Nonclassic Targets in Psychedelic Drug Action. J. Neurosci. 2023, 43, 7472–7482
7. Bepler, T.; Morin, A.; Rapp, M.; et al. TOPAZ: A Positive-Unlabeled Convolutional Neural Network CryoEM Particle Picker that can Pick Any Size and Shape Particle. Microscopy and Microanalysis, 2019, 25 (S2) 986-987.
8. Scheres, S. H.; RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012, 180 (3), 19-30.


